For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SWSRDGMAARVATEFEDGQYINLGIGMSTLIPNHIPEGVTVVLHSENGILGVGPYPIREEFDADLINAGK ETVTTLPGAAFFGSSTSFGIIRGGHLDVAVLGAMQVSVSGDLANWMIPGKMVKGMGGAMDLVHGARKVIV MMEHAAKDGSPKILDRCTLPLTGVGCVNRIVTELAVIDVCHDGLHLIETAPDVSAEEVVAKTQPPLVLDD LVAR*
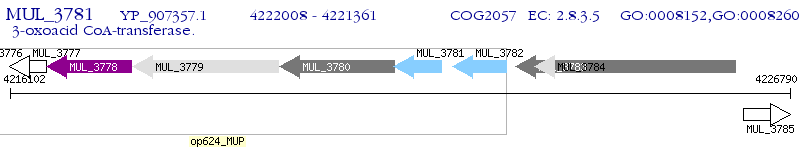
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_3781 | scoB | - | 100% (215) | succinyl-CoA:3-ketoacid-coenzyme a transferase (beta subunit) ScoB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2531c | scoB | 1e-108 | 88.73% (213) | succinyl-CoA:3-ketoacid-coenzyme A transferase subunit beta |
| M. gilvum PYR-GCK | Mflv_1350 | - | 2e-73 | 60.87% (207) | 3-oxoacid CoA-transferase, B subunit |
| M. tuberculosis H37Rv | Rv2503c | scoB | 1e-108 | 88.73% (213) | succinyl-CoA:3-ketoacid-coenzyme A transferase subunit beta |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_3850 | scoB | 1e-120 | 98.60% (215) | succinyl-CoA:3-ketoacid-coenzyme A transferase (beta |
| M. avium 104 | MAV_4786 | - | 9e-68 | 60.00% (200) | succinyl-CoA:3-ketoacid-coenzyme A transferase 1 |
| M. smegmatis MC2 155 | MSMEG_1899 | - | 3e-76 | 62.50% (208) | succinyl-CoA:3-ketoacid-coenzyme A transferase subunit B |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4892 | - | 2e-75 | 66.83% (208) | 3-oxoacid CoA-transferase, B subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1350|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1899|M.smegmatis_MC2_155 --------------------------------------------------
MUL_3781|M.ulcerans_Agy99 --------------------------------------------------
MMAR_3850|M.marinum_M --------------------------------------------------
Mb2531c|M.bovis_AF2122/97 --------------------------------------------------
Rv2503c|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_4892|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_4786|M.avium_104 MSKVHPDAESALHGLLKDGITVAAGGFGLCGIPEKLIQALVDSGIKDLTI
Mflv_1350|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1899|M.smegmatis_MC2_155 --------------------------------------------------
MUL_3781|M.ulcerans_Agy99 --------------------------------------------------
MMAR_3850|M.marinum_M --------------------------------------------------
Mb2531c|M.bovis_AF2122/97 --------------------------------------------------
Rv2503c|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_4892|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_4786|M.avium_104 VGNNAGVDDFGMGLLLKGRQVKKVIASYVGENKEFERQVLSGELDLVLTP
Mflv_1350|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1899|M.smegmatis_MC2_155 --------------------------------------------------
MUL_3781|M.ulcerans_Agy99 --------------------------------------------------
MMAR_3850|M.marinum_M --------------------------------------------------
Mb2531c|M.bovis_AF2122/97 --------------------------------------------------
Rv2503c|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_4892|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_4786|M.avium_104 QGTLAEKLRAGGAGIPGFYTRTGYGTTLAEGKDTRVFDGVEYVLEEGIRA
Mflv_1350|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1899|M.smegmatis_MC2_155 --------------------------------------------------
MUL_3781|M.ulcerans_Agy99 --------------------------------------------------
MMAR_3850|M.marinum_M --------------------------------------------------
Mb2531c|M.bovis_AF2122/97 --------------------------------------------------
Rv2503c|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_4892|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_4786|M.avium_104 DLAIVKAWKGDKAGNLVYRKTARNFNPMIATCAAVTVAEVEELVEVGDLD
Mflv_1350|M.gilvum_PYR-GCK -------------------------------MS------WTRTEMAARAA
MSMEG_1899|M.smegmatis_MC2_155 -------------------------------MS------WNRDDMAARAA
MUL_3781|M.ulcerans_Agy99 -------------------------------MS------WSRDGMAARVA
MMAR_3850|M.marinum_M -------------------------------MS------WSRDGMAARVA
Mb2531c|M.bovis_AF2122/97 -------------------------------MSA---PGWSRDEMAARVA
Rv2503c|M.tuberculosis_H37Rv -------------------------------MSA---PGWSRDEMAARVA
Mvan_4892|M.vanbaalenii_PYR-1 -------------------------------MTATFTTGWTRNQIAARAA
MAV_4786|M.avium_104 PDLIHTPGIYVDRIIQGIGYEKRIEFRTTSGGAALKNDSPIRERMAQRAA
: * :* *.*
Mflv_1350|M.gilvum_PYR-GCK RELRDGDYVNLGIGLPTLIPDHLPEGSDVTLHAENGILGVGPFPYDDEVD
MSMEG_1899|M.smegmatis_MC2_155 TELHDGDYVNLGIGLPTLIPDHLPAGSQVTLHSENGILGVGPFPYDDEVD
MUL_3781|M.ulcerans_Agy99 TEFEDGQYINLGIGMSTLIPNHIPEGVTVVLHSENGILGVGPYPIREEFD
MMAR_3850|M.marinum_M TEFEDGQYINLGIGMPTLIPNHIPEGVTVVLHSENGILGVGPYPIREELD
Mb2531c|M.bovis_AF2122/97 AEFEDGQYVNLGIGMPTLIPNHIPDGVHVVLHSENGILGVGPYPRREDVD
Rv2503c|M.tuberculosis_H37Rv AEFEDGQYVNLGIGMPTLIPNHIPDGVHVVLHSENGILGVGPYPRREDVD
Mvan_4892|M.vanbaalenii_PYR-1 AEMIDGQYVDLGIGLPTLIPGHLSPGVRVILHSENGILGTGGYPDEDSVD
MAV_4786|M.avium_104 KELRDGYYVNLGIGIPTLVANFIPDDIQVTLQSENGLLGIGAYPSEDTVD
*: ** *::****:.**:...:. . * *::***:** * :* : .*
Mflv_1350|M.gilvum_PYR-GCK PDLVNAGKQTVSVVAGASFFDSATSFAMIRGGHVDVAVLGGMQVAVNGDL
MSMEG_1899|M.smegmatis_MC2_155 PDLVNAGKQTVSVVAGASYFDSATSFAMIRGGHVDVAVLGGMQVAANGDL
MUL_3781|M.ulcerans_Agy99 ADLINAGKETVTTLPGAAFFGSSTSFGIIRGGHLDVAVLGAMQVSVSGDL
MMAR_3850|M.marinum_M ADLINAGKETVTTLPGAAFFGSSTSFGIIRGGHLDVAVLGAMQVSVSGDL
Mb2531c|M.bovis_AF2122/97 ADLINAGKETVTTLPGAAFFSSSTSFGIIRGGHLDVAVLGAMQVSVTGDL
Rv2503c|M.tuberculosis_H37Rv ADLINAGKETVTTLPGAAFFSSSTSFGIIRGGHLDVAVLGAMQVSVTGDL
Mvan_4892|M.vanbaalenii_PYR-1 ADLINAGKETVTVNRGAAFFDSATSFGMIRGGHIDAAVLGAMQVSRNGDL
MAV_4786|M.avium_104 PDLINAGKQTITSLPGSSFFSSADSFAMIRGGHIDLSILGALEVASNGDL
.**:****:*:: *:::*.*: **.:*****:* ::**.::*: .***
Mflv_1350|M.gilvum_PYR-GCK ANWMVPGSMVKGMGGAMDLVNGAAKVIVLMDHVSKNGAAKLVSSCDLPLT
MSMEG_1899|M.smegmatis_MC2_155 ANWMVPGSMVKGMGGAMDLVSGARRVIVLMDHVAKSGAAKLVSSCTLPLT
MUL_3781|M.ulcerans_Agy99 ANWMIPGKMVKGMGGAMDLVHGARKVIVMMEHAAKDGSPKILDRCTLPLT
MMAR_3850|M.marinum_M ANWMIPGKMVKGMGGAMDLVHGARKVIVMMEHAAKDGSPKILERCTLPLT
Mb2531c|M.bovis_AF2122/97 ANWMIPGKMVKGMGGAMDLVHGARKVIVMMEHTAKDGSPKILERCTLPLT
Rv2503c|M.tuberculosis_H37Rv ANWMIPGKMVKGMGGAMDLVHGARKVIVMMEHTAKDGSPKILERCTLPLT
Mvan_4892|M.vanbaalenii_PYR-1 ANWMVPGKMVKGMGGAMDLVHGAARVIVVMEHTDRSGAPKIVDACTLPLT
MAV_4786|M.avium_104 ANWMVPGKMVKGPGGAMDLVSGVKRVIVLMDHTAKDGSPKVLEQCTLPLT
****:**.**** ******* *. :***:*:*. :.*:.*::. * ****
Mflv_1350|M.gilvum_PYR-GCK GKAVVQRVITDLGVFDVTGTE-FGVVELSPGVDLDEVAAKTGAPVSDHRH
MSMEG_1899|M.smegmatis_MC2_155 GKAVVSRVITDLGVFDVTGDG-FRVVELAPGVEFDHVAANTDATLVDG--
MUL_3781|M.ulcerans_Agy99 GVGCVNRIVTELAVIDVCHDG-LHLIETAPDVSAEEVVAKTQPPLVLD--
MMAR_3850|M.marinum_M GVGCVNRIVTELAVIDVCHDG-LHLIETAPDVSAEEVVAKTQPPLVLD--
Mb2531c|M.bovis_AF2122/97 GVGCVDRIVTELAVIDVCADG-LHLVQTAPGVSVDEVVAKTQPPLVLR--
Rv2503c|M.tuberculosis_H37Rv GVGCVDRIVTELAVIDVCADG-LHLVQTAPGVSVDEVVAKTQPPLVLR--
Mvan_4892|M.vanbaalenii_PYR-1 GQGVVDRIITNLCVIDVPGDGTLVLRELAPGVTVDDVVAATGAELTVA--
MAV_4786|M.avium_104 GKGVVDMIITNLCVFDVEPGRGLLLTELHPGVTVEEVRAKTGCDFTVAL-
* . *. ::*:* *:** : : : *.* :.* * * .
Mflv_1350|M.gilvum_PYR-GCK RLSAASSERDRMPSLA
MSMEG_1899|M.smegmatis_MC2_155 ----------RVGSLV
MUL_3781|M.ulcerans_Agy99 ----------DLVAR-
MMAR_3850|M.marinum_M ----------DLVAR-
Mb2531c|M.bovis_AF2122/97 ----------DLATQ-
Rv2503c|M.tuberculosis_H37Rv ----------DLATQ-
Mvan_4892|M.vanbaalenii_PYR-1 ----------L-----
MAV_4786|M.avium_104 ----------------