For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSTARPLRVIQWTTGNIGRRSLHAIIGRPDLELVGVYAHGPDKVGVDAAELAGWPDPTGVRATSDIGALL ALGADACCYNPLWPNIDELVGLLESGVNVCTSAAWITGGKQTPRDRERILAACERGGATIFGSDAHPGMT NMVGMVLSGSCERVDEIRITESVDCSTYESAATQTAMGFSQHPDTPGLAESVRVESEVFAESAAMMADAI GATLDKMTFDVTFTAATADTDLGFMKIPAGTVGSVYGYHRGWVGDRNVVSVGFNWTMGNHVVPPKPLKHG HVIQVFGLPNMRTVLHCLPPKDWTESGFMGLGMIYTAMPVTNAVPAVVAAKPGIVTLADLPPITGRAAL
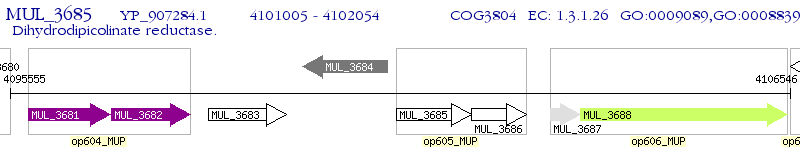
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_3685 | - | - | 100% (349) | hypothetical protein MUL_3685 |
| M. ulcerans Agy99 | MUL_2162 | - | 1e-54 | 35.89% (326) | hypothetical protein MUL_2162 |
| M. ulcerans Agy99 | MUL_4353 | - | 2e-37 | 31.84% (358) | hypothetical protein MUL_4353 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1088 | - | 2e-41 | 33.05% (351) | hypothetical protein Mb1088 |
| M. gilvum PYR-GCK | Mflv_2658 | - | 1e-175 | 83.62% (348) | dihydrodipicolinate reductase |
| M. tuberculosis H37Rv | Rv1059 | - | 2e-41 | 33.05% (351) | hypothetical protein Rv1059 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0727 | - | 3e-54 | 34.29% (347) | putative dihydrodipicolinate reductase |
| M. marinum M | MMAR_3741 | - | 0.0 | 98.85% (349) | hypothetical protein MMAR_3741 |
| M. avium 104 | MAV_1757 | dapB | 0.0 | 90.72% (345) | dihydrodipicolinate reductase N-terminus domain-containing |
| M. smegmatis MC2 155 | MSMEG_0560 | dapB | 2e-76 | 46.24% (346) | dihydrodipicolinate reductase N-terminus domain-containing |
| M. thermoresistible (build 8) | TH_4249 | - | 3e-74 | 43.70% (341) | PUTATIVE dihydrodipicolinate reductase |
| M. vanbaalenii PYR-1 | Mvan_3920 | - | 1e-176 | 84.93% (345) | dihydrodipicolinate reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1088|M.bovis_AF2122/97 ---------------MTMSLRVIQWATGSVGVAAIKGVLQHPELELVGCW
Rv1059|M.tuberculosis_H37Rv ---------------MTMSLRVIQWATGSVGVAAIKGVLQHPELELVGCW
MUL_3685|M.ulcerans_Agy99 ----------MS---TARPLRVIQWTTGNIGRRSLHAIIGRPDLELVGVY
MMAR_3741|M.marinum_M ----------MS---TERPLRVIQWTTGNIGRRSLHAIIGRPDLELVGVY
MAV_1757|M.avium_104 MRQSHGKGAPMSKPSTTRPLRVIQWTTGNIGRRSLHAIIGRPDMELVGVY
Mflv_2658|M.gilvum_PYR-GCK --------MPPTTT-TDRPLRVIQWTTGNIGRRSLHAIIGRDDMELVGVF
Mvan_3920|M.vanbaalenii_PYR-1 ----------MATP-NERPLRVIQWTTGNIGRRSLHAIIGRDDMELVGVF
MSMEG_0560|M.smegmatis_MC2_155 -------------MPDTTGYRVVQWTTGNVGKSSVAAIAKNPTLELVGCY
TH_4249|M.thermoresistible__bu -------------VPNTP-YRVVQWTTGNVGKSSVQALAANPLYELVGVY
MAB_0727|M.abscessus_ATCC_1997 ----------------MSVIRVFQVATGNVGTEMIRRIASHPDLELAGLH
**.* :**.:* : : . **.*
Mb1088|M.bovis_AF2122/97 VHSAAKSGKDVGEIIGSP-PLGVIATNSIDDVLALDADAVIYAPLLPSVD
Rv1059|M.tuberculosis_H37Rv VHSAAKSGKDVGEIIGSP-PLGVIATNSIDDVLALDADAVIYAPLLPSVD
MUL_3685|M.ulcerans_Agy99 AHGPDKVGVDAAELAGWPDPTGVRATSDIGALLALGADACCYNPLWPNID
MMAR_3741|M.marinum_M AHGPDKVGVDAAELAGWPDPTGVRATSDIGALLALGADACCYNPLWPNID
MAV_1757|M.avium_104 AHGAEKVGVDAAELAGWPEPTGVAATNDIDALLSLGADACCYNPLWPNVD
Mflv_2658|M.gilvum_PYR-GCK AHGADKVGVDAADLAGRPEPTGVKATDDIEELVALRPDACCYNPLWPSID
Mvan_3920|M.vanbaalenii_PYR-1 AHGPDKVGVDAAELAGWPEPTGVTATDDIEELVALKPDACCYNPLWPSVD
MSMEG_0560|M.smegmatis_MC2_155 AWSGDKVGKDVGELCGIE-PLGVSATDDVDALLALKPDVVVYNPMWIDVD
TH_4249|M.thermoresistible__bu AWSPDKVGRDAGELAGIE-PLGVKATNDVDELLALKPDVVVYNPMWIDVD
MAB_0727|M.abscessus_ATCC_1997 CYSPEKVGRDAGELVGLE-PNGVIATGTVEEIVAAKPDVLTFHGVFPDED
. * * *..:: * * ** **. : ::: .*. : : . *
Mb1088|M.bovis_AF2122/97 EVAALLRSGKNVVTPLGWFYP---SEKEAAP--------LEVAAQAGNAT
Rv1059|M.tuberculosis_H37Rv EVAALLRSGKNVVTPLGWFYP---SEKEAAP--------LEVAAQAGNAT
MUL_3685|M.ulcerans_Agy99 ELVGLLESGVNVCTSAAWITGGKQTPRDRER--------ILAACERGGAT
MMAR_3741|M.marinum_M ELVGLLESGVNVCTSAAWITGGKQTPRDRER--------ILAACERGGAT
MAV_1757|M.avium_104 ELVRLLESGVNVCTSAAWITGGKQTPEDRRR--------IEEACRKGNST
Mflv_2658|M.gilvum_PYR-GCK ELVRLLESGINVCSSAAWITGGKQSPEDLDR--------IRKACETGNST
Mvan_3920|M.vanbaalenii_PYR-1 ELVRLLESGVNVCSSAAWITGAKQSPEDLDR--------IRKACATGNST
MSMEG_0560|M.smegmatis_MC2_155 ELVRILESGVNVVSTASFITG-HNQGTGRDR--------IAEACTRGGSS
TH_4249|M.thermoresistible__bu ELVRILSAGINVVSSASFITG-HNLRDGRDR--------IEEACRQGGST
MAB_0727|M.abscessus_ATCC_1997 LYVKVLEAGIDIVTTADWITGHHRDQNHPHPSGRPTTEVLQQACERGGST
. :* :* :: :. :: : *. *.::
Mb1088|M.bovis_AF2122/97 LHGAGIGPGAVTELFPLLLSVMSTGVTFVRSEEFSDLRSYGAPDVLRYVM
Rv1059|M.tuberculosis_H37Rv LHGAGIGPGAVTELFPLLLSVMSTGVTFVRSEEFSDLRSYGAPDVLRYVM
MUL_3685|M.ulcerans_Agy99 IFGSDAHPG-MTNMVGMVLSGSCERVDEIRITESVDCSTYES-AATQTAM
MMAR_3741|M.marinum_M IFGSGAHPG-MTNMVGMVLSGSCERVDEIRITESVDCSTYES-AATQTAM
MAV_1757|M.avium_104 IFGSGAHPG-MTNMVGMVLSASCERVDEIRITESVDCSTYES-AETQTAM
Mflv_2658|M.gilvum_PYR-GCK IFGSGAHPG-MSNLVGMVLSGACERVDEIRITESVDCSTYES-AGTQTAM
Mvan_3920|M.vanbaalenii_PYR-1 IFGSGAHPG-MSNLVGMVLSGACERVDEIRITESVDCSTYES-AGTQTAM
MSMEG_0560|M.smegmatis_MC2_155 MFGSGISPG-YVNQLSVVAAGICDRVDKITVNEAADTTFYDS-PATEKPV
TH_4249|M.thermoresistible__bu MFGSGISPG-FAELLAIIGTQMCDRVDKVTICEQSDTTFYDS-PATEKPV
MAB_0727|M.abscessus_ATCC_1997 FYGTGMNPG-LTQILGVIHSCDVAEIENVTVIESVDVSCHHS-VDSWEMV
:.*:. ** : . :: : : : * * : : :
Mb1088|M.bovis_AF2122/97 GFGGTPDSALTGPMQKILDGGFLQSVRLCVDRLGFAADPQIRTSQEVAVA
Rv1059|M.tuberculosis_H37Rv GFGGTPDSALTGPMQKILDGGFLQSVRLCVDRLGFAADPQIRTSQEVAVA
MUL_3685|M.ulcerans_Agy99 GFSQHPDTPGLAESVRVESEVFAESAAMMADAIGATLD-KMTFDVTFTAA
MMAR_3741|M.marinum_M GFSQHPDTPGLAESVRVESEVFAESAAMMADAIGATLD-KMTFDVTFTAA
MAV_1757|M.avium_104 GFSQDPDTPGLAESVRRESEVFAESAAMMADAIGAKLD-KMTFDVTFTAA
Mflv_2658|M.gilvum_PYR-GCK GFSQDPDTPGLAESVRRESEVFAESAAMMADAIGATLD-RITFDVTFTAA
Mvan_3920|M.vanbaalenii_PYR-1 GFSQDPDTPGLAESVRRESEVFAESAAMMADAIGARLD-RITFDVTFTAA
MSMEG_0560|M.smegmatis_MC2_155 GFGQPIDHPDLQAMTAQGTGVFGEAVRMIADALGIELD-EVRCDAEYAQT
TH_4249|M.thermoresistible__bu GFGTPIDDPNLPQMASQGTAIFAEAVQLIADSLGVELD-EIVCEAEYAQT
MAB_0727|M.abscessus_ATCC_1997 GYGLPVGDPSIPALLEKGTRVFADGVYLMADCFGLELD-DVTFSYELGAC
*:. . . * :.. : .* :* * : .
Mb1088|M.bovis_AF2122/97 TAPIDSPIGVIEPGQVAGRRFHWEALVEDTVVVQIAVNWLMGSENLDPPW
Rv1059|M.tuberculosis_H37Rv TAPIDSPIGVIEPGQVAGRRFHWEALVEDTVVVQIAVNWLMGSENLDPPW
MUL_3685|M.ulcerans_Agy99 TADTDLGFMKIPAGTVGSVYGYHRGWVGDRNVVSVGFNWTMGNHVVPP--
MMAR_3741|M.marinum_M TADTDLGFMKIPAGTVGSVYGYHRGWVGDRNVVSVGFNWTMGNHVVPP--
MAV_1757|M.avium_104 TGDSDLGFMKIPAGTVGSVYGYHRGWVGDRNVVSVGFNWTMGEHVVPP--
Mflv_2658|M.gilvum_PYR-GCK TADSDLGFMQIPKGTVAGVHGYHRGWVGDHNIVSVGFNWIMGEHVTPP--
Mvan_3920|M.vanbaalenii_PYR-1 TGDSDLGFMQIPAGTVAGVHGYHRGWVGDHNIVSVGFNWTMGEHVTPP--
MSMEG_0560|M.smegmatis_MC2_155 TEDLDLGSWTIPAGCVAGVYASWKGIVAGHTRVEINVRWRKGQTLQPD--
TH_4249|M.thermoresistible__bu TEDLDLGSWKIGAGCVAGVYASWQGRIGGRTLIDLNVRWKKGQTLEPN--
MAB_0727|M.abscessus_ATCC_1997 TEDIDLGWYRLPKGSLGANYLKYQGIVGGVPKVEVHVEWQMTPKTEPH--
* * : * :.. .. : . :.: ..*
Mb1088|M.bovis_AF2122/97 SFGPAGERYEIEVRGSPDTCVTIKGWQPQTVAAG---LKSNPGIVATAAH
Rv1059|M.tuberculosis_H37Rv SFGPAGERYEIEVRGSPDTCVTIKGWQPQTVAAG---LKSNPGIVATAAH
MUL_3685|M.ulcerans_Agy99 --KPLKHGHVIQVFGLPNMRTVLHCLPPKDWTES---GFMGLGMIYTAMP
MMAR_3741|M.marinum_M --KPLEHGHVIQVFGLPNMRTVLHCLPPKDWTEP---GFMGLGMIYTAMP
MAV_1757|M.avium_104 --KPLEHGHVIQVFGLPNMRTVLHCLPPKDWTEP---GFMGLGMIYTAMP
Mflv_2658|M.gilvum_PYR-GCK --KPLEHGHVVQVFGLPNMRTVVHCLPPRDWTEP---GFMGLGMIYTAMP
Mvan_3920|M.vanbaalenii_PYR-1 --KPLEHGHVVQVFGLPNMRTVLHCLPPKDWTEP---GFMGLGMIYTAMP
MSMEG_0560|M.smegmatis_MC2_155 -WKIDQDGWVIEVAGRPTVTMKVGFLPPPDFEATTLEEFMVLGHIMTATP
TH_4249|M.thermoresistible__bu -WQLEREGWKITVDGRPTVNMEIGFLPPPDFGATTMEEFMVLGHIMTAMP
MAB_0727|M.abscessus_ATCC_1997 --WNVKGCYITKIQGDPCVYSKHMVIPKPGTDFTDPAAFASVGMTVTGLP
: * * * *.
Mb1088|M.bovis_AF2122/97 CVNAIPATCAAPAGIQSFFDLPLITGRAAPGLAR
Rv1059|M.tuberculosis_H37Rv CVNAIPATCAAPAGIQSFFDLPLITGRAAPGLAR
MUL_3685|M.ulcerans_Agy99 VTNAVPAVVAAKPGIVTLADLPPITGRAAL----
MMAR_3741|M.marinum_M VTNAVPAVVAAKPGIVTLADLPPITGRAAL----
MAV_1757|M.avium_104 VTNAVPAVVAARPGIVTLADLPPVTGRLG-----
Mflv_2658|M.gilvum_PYR-GCK VTNAVPAVVAAPPGIVTLADLPPITGRSAV----
Mvan_3920|M.vanbaalenii_PYR-1 VTNAVPAVVAAPPGIVTLADLPPITGRSAV----
MSMEG_0560|M.smegmatis_MC2_155 PLNAIPAVVAAPPGIVTYNDLPLILPRGVVPTP-
TH_4249|M.thermoresistible__bu PIHMIPAVVAAEPGIATYNDLPLPQARGVVRV--
MAB_0727|M.abscessus_ATCC_1997 ALNAIRSVIDAPPGILTSADLPLRGFAGRFK---
: : :. * .** : ***