For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SMLRVAVPNKGALSEPATEILAEAGYRRRTDPKDLTVIDPANNVEFFFLRPKDIAIYVGSGQLDFGITGR DLVHDSDAQVRERLALGFGSSSFRYAGPAQRDWTMADLAGKRIATAYPNLVRKDLTAKGIEATVIRLDGA VEISVQLGVANAIADVVGSGRTLRLHNLVAFGEPLCDSEAVLIEGNGRDGAAQVEARDQLVARVQGVVFG QQYLMLDYDCPRAVLDKATLITPGLESPTIAPLADPEWVAVRALVPRRDVNGTMDELAAIGAKAILASDI RFCRF*
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
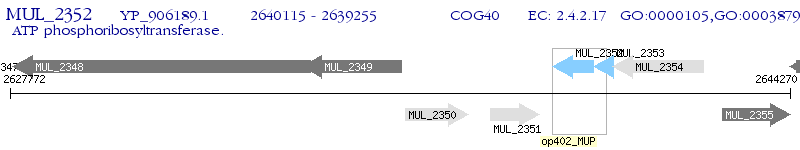
| M. ulcerans Agy99 | MUL_2352 | hisG | - | 100% (286) | ATP phosphoribosyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2145c | hisG | 1e-141 | 89.79% (284) | ATP phosphoribosyltransferase |
| M. gilvum PYR-GCK | Mflv_3057 | hisG | 1e-132 | 83.80% (284) | ATP phosphoribosyltransferase |
| M. tuberculosis H37Rv | Rv2121c | hisG | 1e-141 | 89.79% (284) | ATP phosphoribosyltransferase |
| M. leprae Br4923 | MLBr_01310 | hisG | 1e-141 | 88.58% (289) | ATP phosphoribosyltransferase |
| M. abscessus ATCC 19977 | MAB_2132 | hisG | 1e-123 | 79.08% (282) | ATP phosphoribosyltransferase |
| M. marinum M | MMAR_3103 | hisG | 1e-160 | 99.30% (286) | ATP phosphoribosyltransferase HisG |
| M. avium 104 | MAV_2393 | hisG | 1e-138 | 88.03% (284) | ATP phosphoribosyltransferase |
| M. smegmatis MC2 155 | MSMEG_4180 | hisG | 1e-136 | 84.86% (284) | ATP phosphoribosyltransferase |
| M. thermoresistible (build 8) | TH_0037 | hisG | 1e-128 | 81.34% (284) | Probable ATP phosphoribosyltransferase HisG |
| M. vanbaalenii PYR-1 | Mvan_3470 | hisG | 1e-129 | 82.69% (283) | ATP phosphoribosyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_2352|M.ulcerans_Agy99 ----MSMLRVAVPNKGALSEPATEILAEAGYRRRTDPKDLTVIDPANNVE
MMAR_3103|M.marinum_M ----MSMLRVAVPNKGALSEPATEILAEAGYRRRTDPKDLTVIDPANNVE
MLBr_01310|M.leprae_Br4923 ----MSMLRVAVPNKGALSEPATEILAEAGYRRRTDPKDLTVVDPVNRVE
Mb2145c|M.bovis_AF2122/97 ------MLRVAVPNKGALSEPATEILAEAGYRRRTDSKDLTVIDPVNNVE
Rv2121c|M.tuberculosis_H37Rv ------MLRVAVPNKGALSEPATEILAEAGYRRRTDSKDLTVIDPVNNVE
MAV_2393|M.avium_104 ------MLRVAVPNKGTLSEPATEILAEAGYRRRTDSKDLTVIDPVNQVE
MSMEG_4180|M.smegmatis_MC2_155 --MTDQMLRVAVPNKGALSESAAEILAEAGYRRRRDPKDLTVVDPGNNVE
TH_0037|M.thermoresistible__bu -----VMLRIAMPNKGTLSEPAAEILAEAGYRRRSDTKDLTVVDPVNEVE
Mflv_3057|M.gilvum_PYR-GCK ------MLRVAVPNKGALSESASEILSEAGYRRRSDPKDLTVVDPANNVE
Mvan_3470|M.vanbaalenii_PYR-1 -MSAPKTLRVAVPNKGALSESAAEILSEAGYRRRRDSKDLTVVDPSNNVE
MAB_2132|M.abscessus_ATCC_1997 MTSTNGALRVAVPNKGALSEAASEILSEAGYRRRGDAKDLTVLDPQNDVE
**:*:****:***.*:***:******* *.*****:** * **
MUL_2352|M.ulcerans_Agy99 FFFLRPKDIAIYVGSGQLDFGITGRDLVHDSDAQVRERLALGFGSSSFRY
MMAR_3103|M.marinum_M FFFLRPKDIAIYVGSGQLDFGITGRDLVHDSDAQVRERLALGFGSSSFRY
MLBr_01310|M.leprae_Br4923 FFFLRPKDIAIYVGSGDLDFGITGRDLVHDSDASVCERLALGFGSSSFRY
Mb2145c|M.bovis_AF2122/97 FFFLRPKDIAIYVGSGELDFGITGRDLVCDSGAQVRERLALGFGSSSFRY
Rv2121c|M.tuberculosis_H37Rv FFFLRPKDIAIYVGSGELDFGITGRDLVCDSGAQVRERLALGFGSSSFRY
MAV_2393|M.avium_104 FFFLRPKDIAIYVGSGELDFGITGRDLVGDSDAPVRERLALGFGSSSFRY
MSMEG_4180|M.smegmatis_MC2_155 FFFLRPKDIAIYVGSGQLDLGITGRDLAADSDAPVRERLSLGFGNSTFRY
TH_0037|M.thermoresistible__bu FFFLRPKDIAIYVGSGDLDLGITGRDLTAESGAPVRERLALGFGSSTFRY
Mflv_3057|M.gilvum_PYR-GCK FFFLRPKDIAIYVGSGELDLGITGRDLAADADAPVRERLALGFGSSTFRY
Mvan_3470|M.vanbaalenii_PYR-1 FFFLRPKDIAIYVGSGELDLGITGRDLAQESDAPVAERLALGFGSSTFRY
MAB_2132|M.abscessus_ATCC_1997 FFFLRPKDIAIYVGSGELDLGITGRDLMMDSDAPVTERLALGFGGSTFRY
****************:**:******* ::.* * ***:****.*:***
MUL_2352|M.ulcerans_Agy99 AGPAQRDWTMADLAGKRIATAYPNLVRKDLTAKGIEATVIRLDGAVEISV
MMAR_3103|M.marinum_M AGPAQRDWTMADLAGKRIATAYPNLVRKDLAAKGIEATVIRLDGAVEISV
MLBr_01310|M.leprae_Br4923 AGPAGRDWTTSDLAGKRIATAYSNLVRKDLVAKGIEATVIRLDGAVEISV
Mb2145c|M.bovis_AF2122/97 AAPAGRNWTTADLAGMRIATAYPNLVRKDLATKGIEATVIRLDGAVEISV
Rv2121c|M.tuberculosis_H37Rv AAPAGRNWTTADLAGMRIATAYPNLVRKDLATKGIEATVIRLDGAVEISV
MAV_2393|M.avium_104 AGPAGRDWKIADLAGKRIATAYPNLVRKDLAERGIEATVIRLDGAVEISV
MSMEG_4180|M.smegmatis_MC2_155 AAPAGREWTTADLAGKRIATSFPNLVRKDLAAHGIDATVIRLDGAVEISI
TH_0037|M.thermoresistible__bu AAPAGRKWTVADLAGKRIATSFPNLVRKDLAARGIDATVIRLDGAVEISV
Mflv_3057|M.gilvum_PYR-GCK AAPAGQDWQIADLAGRRIATAYPNLVRKDLADKGIDATVIRLDGAVEISI
Mvan_3470|M.vanbaalenii_PYR-1 AAPAGTDWTVADLAGRRIATAYPNLVRKDLAHRGIEATVIRLDGAVEISV
MAB_2132|M.abscessus_ATCC_1997 AAPAGRDWTIYDIAGKRIATAYPNLVRKDLAAKGIEAEVIRLDGAVEISI
*.** .* *:** ****::.*******. :**:* ***********:
MUL_2352|M.ulcerans_Agy99 QLGVANAIADVVGSGRTLRLHNLVAFGEPLCDSEAVLIEGNGR---DGAA
MMAR_3103|M.marinum_M QLGVADAIADVVGSGRTLRLHNLVAFGEPLCDSEAVLIEGNGR---DGAA
MLBr_01310|M.leprae_Br4923 QLGVADVIADVVGSGRTLSLHDLVAFGEPLCDSEAVLIERGDRGSLDHHE
Mb2145c|M.bovis_AF2122/97 QLGVADAIADVVGSGRTLSQHDLVAFGEPLCDSEAVLIERAGT---DGQD
Rv2121c|M.tuberculosis_H37Rv QLGVADAIADVVGSGRTLSQHDLVAFGEPLCDSEAVLIERAGT---DGQD
MAV_2393|M.avium_104 QLGVADAIADVVGSGRTLSLHNLVAFGEPLCDSEAVLIESDRG---GQDD
MSMEG_4180|M.smegmatis_MC2_155 QLGVADVIADVVGSGRTLGLHNLVAFGDPLCESEAVLIERDGADG---DE
TH_0037|M.thermoresistible__bu QLGVADAIADVVGSGRTLGLHGLVAFGESLCDSEAVLIERIDAD----PS
Mflv_3057|M.gilvum_PYR-GCK QLGVADVIADIVGSGRTLGLHNLVAFGESLCDSEAILIERADS----DPD
Mvan_3470|M.vanbaalenii_PYR-1 QLGLADVIADIVGTGRTLGLHGLVAFGEALCDSEAVLIERVDA----DPA
MAB_2132|M.abscessus_ATCC_1997 QLGVADVIADIVGTGRTLRQHGLVAFGESLCDSEAVLIERENA----DPA
***:*:.***:**:**** *.*****:.**:***:***
MUL_2352|M.ulcerans_Agy99 QVEARDQLVARVQGVVFGQQYLMLDYDCPRAVLDKATLITPGLESPTIAP
MMAR_3103|M.marinum_M QVEARDQLVARVQGVVFGQQYLMLDYDCPRAVLDKATLITPGLESPTIAP
MLBr_01310|M.leprae_Br4923 TVAARDQLVARVQGVVFGQQYLMLDYDCPRSVLDKATLITSGLESPTIAP
Mb2145c|M.bovis_AF2122/97 QTEARDQLVARVQGVVFGQQYLMLDYDCPRSALKKATAITPGLESPTIAP
Rv2121c|M.tuberculosis_H37Rv QTEARDQLVARVQGVVFGQQYLMLDYDCPRSALKKATAITPGLESPTIAP
MAV_2393|M.avium_104 TQAARDQLVARIQGVVFGQQYLMLDYDCPRAVLDKATAITPGLESPTIAP
MSMEG_4180|M.smegmatis_MC2_155 NAAARDQLTARVQGVVFGQQYLMLDYDCPRAVLDRATAVTPGLESPTIAP
TH_0037|M.thermoresistible__bu NAAARDKLVARVQGVVFGQQYLMLDYDCPRDVLDDAMAITPGLQSPTVAP
Mflv_3057|M.gilvum_PYR-GCK --PARDQLAARVQGVVFGQQYLMLDYDCPRTVLDKATEVTPGLESPTIAP
Mvan_3470|M.vanbaalenii_PYR-1 TAAARDQLAARVQGVVFGQQYLMLDYDCPRSVLERATEVTPGLESPTIAP
MAB_2132|M.abscessus_ATCC_1997 TESARTQLTTRIQGVVFGQQYLMLDYDCPRAVLDEALKVTPGLESPTLAP
** :*.:*:****************** .*. * :*.**:***:**
MUL_2352|M.ulcerans_Agy99 LADPEWVAVRALVPRRDVNGTMDELAAIGAKAILASDIRFCRF
MMAR_3103|M.marinum_M LADPEWVAVRALVPRRDVNGTMDELAAIGAKAILASDIRFCRF
MLBr_01310|M.leprae_Br4923 LAEPGWVAIRALVPRRDINGIMDELAAIGAKAILASDIRFCRF
Mb2145c|M.bovis_AF2122/97 LADPDWVAIRALVPRRDVNGIMDELAAIGAKAILASDIRFCRF
Rv2121c|M.tuberculosis_H37Rv LADPDWVAIRALVPRRDVNGIMDELAAIGAKAILASDIRFCRF
MAV_2393|M.avium_104 LADADWVAIRALVPRRGVNEIMDELAAIGAKAILASDIRFCRF
MSMEG_4180|M.smegmatis_MC2_155 LADPDWVAVRALVPRRDVNAIMDEIAAIGAKAILASDIRFCRF
TH_0037|M.thermoresistible__bu LADPAWVAVRAMVPRRDVNAIMDELGAIGAKAILASEIRFCRF
Mflv_3057|M.gilvum_PYR-GCK LADPAWVAVRALVPRRDVNAIMDELAAIGAKAILASDIRFCRF
Mvan_3470|M.vanbaalenii_PYR-1 LADPDWVAVRALVPRRDVNAIMDELAAIGAKAILASDIRFCRF
MAB_2132|M.abscessus_ATCC_1997 LADEAWVAVRALAPRKGVNTTMDALAAIGAKAILASEVRFFRA
**: ***:**:.**:.:* ** :.**********::** *