For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVGTVGAMGRVQAAGDNAAMASFFSLLRKNVLGRRRWETREHLRIAIATWTETHLRPPPSASRSWPVAPV RFEAIMISRAIQAA
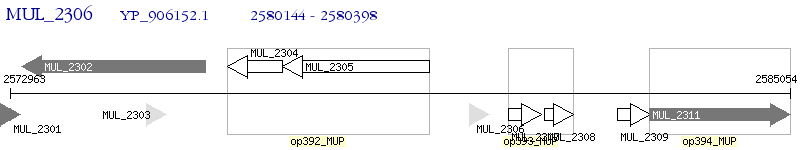
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_2306 | - | - | 100% (84) | transposase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3195 | - | 4e-21 | 62.50% (80) | integrase catalytic subunit |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2088 | - | 6e-21 | 60.98% (82) | transposase |
| M. marinum M | MMAR_3786 | - | 9e-06 | 31.58% (76) | transposase |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_6888 | - | 4e-20 | 60.00% (80) | transposase subunit |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5959 | - | 1e-20 | 62.50% (80) | integrase catalytic subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3195|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5959|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6888|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2088|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_2306|M.ulcerans_Agy99 --------------------------------------------------
MMAR_3786|M.marinum_M MPRQYSSSVRRQIVSRLRVGEPVAVVAAETGVCQATLFRWKRQALIDAGV
Mflv_3195|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5959|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6888|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2088|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_2306|M.ulcerans_Agy99 --------------------------------------------------
MMAR_3786|M.marinum_M IEGTPSVEADELAAAHKRIAQLEAELALTRDACDLFNEEAVVPPKRRRAI
Mflv_3195|M.gilvum_PYR-GCK MKELAADGIPVAVTCRVLKLSRQPYYRWLADPITEAELIEAYRANALFDA
Mvan_5959|M.vanbaalenii_PYR-1 MKELAADGIPVAVTCRVLKLSRQPYYRWLADPVTDAELVEAYRTNALVNA
MSMEG_6888|M.smegmatis_MC2_155 ------------MTCRVLKLARQPYYRWRANPITDAEIIEAYRANALFDA
MAB_2088|M.abscessus_ATCC_1997 ------------MTRRVLKLARQPYYRWRANPITDAELVEAYRANALFDA
MUL_2306|M.ulcerans_Agy99 --------------------------------------------------
MMAR_3786|M.marinum_M TEGLIARGHSARSACRVTGLARSLLQYHRRRLVPDREVRRLIVADTITEI
Mflv_3195|M.gilvum_PYR-GCK HADDPEFGYRYLVEEARDAGEPMAERTAWR--ICSQNRLWSVFGKKRGKN
Mvan_5959|M.vanbaalenii_PYR-1 HADDPEFGYRYLVEEAREAGEPMAERTAWR--ICSQNRLWSVFGKKRGKS
MSMEG_6888|M.smegmatis_MC2_155 HRDDPEFGYRYLVEEARDAGETMAERTAWR--ICSQNRLWSVFGKKRGKN
MAB_2088|M.abscessus_ATCC_1997 HGEDPEFGYRYLVEEAREAGEPMAERTAWR--ICSQQQLWSVFGKKRGKN
MUL_2306|M.ulcerans_Agy99 --------------------------------------------------
MMAR_3786|M.marinum_M YQRSRGTYGRKRVRAALLADYDMNVNHKLVNSIMSEHGLYGLPRHGRRKP
Mflv_3195|M.gilvum_PYR-GCK GKVGPPVHDDLVERDFTAEGPNQLWLSDITEHRTGEGKLYLCAIKDVFSN
Mvan_5959|M.vanbaalenii_PYR-1 GKVGPPVHDDLVQRDFTADAPNQLWLSDITEHRTDEGKLYLCAIKDVFSN
MSMEG_6888|M.smegmatis_MC2_155 GKVGPPVHDDLVERDFTAGGPNQLWLSDITEHRTDEGKLYLCAIKDVFSN
MAB_2088|M.abscessus_ATCC_1997 GKPGPPVHDDLVQRDFTAETPNMLWLSDITEHYTGDGKLYLCAIKDAFSN
MUL_2306|M.ulcerans_Agy99 --------------------------------------------------
MMAR_3786|M.marinum_M NLIGVHTPVDLINRRFTATKPNELWCTDITEHPARDGKVYCCAILDCFSR
Mflv_3195|M.gilvum_PYR-GCK RIVGYSIDSRMKSQLAIRALHSAVARRGDVAGCILHSDRGSQFRSRKFVH
Mvan_5959|M.vanbaalenii_PYR-1 RIVGYSIDSRMKSRLATRALHSAVARRGDVAGCIVHTDRGSQFRSRRFVH
MSMEG_6888|M.smegmatis_MC2_155 RIVGYSIDARMKSRLATTALSSAVARRGDVAGCILHSDRGSQFRSRRFVR
MAB_2088|M.abscessus_ATCC_1997 RIVGYSIDSRMKSRLATAALRNAVGRRGVVAGCVVHTDRGSQFRSRKFVQ
MUL_2306|M.ulcerans_Agy99 -MVG----------------------------------------------
MMAR_3786|M.marinum_M MIVARTFSTTADTALVNNAVNMAVDHRNRSGPTILHADHGTQFTSWSFGE
:*.
Mflv_3195|M.gilvum_PYR-GCK ALNHHEMVGSMGRVGAAGDNAAMESFFSLLQKNVLDRRRWDTREQLRIAI
Mvan_5959|M.vanbaalenii_PYR-1 ALGHYEMVGSMGRVGAAGDNAAMESFFSLLQKNVLDRRRWATREELRIAI
MSMEG_6888|M.smegmatis_MC2_155 VLGRHEMVGSMGRVGAAGDNAAMESFFALLQKNVLDRRRWRTREELRIAI
MAB_2088|M.abscessus_ATCC_1997 ALHHHGMVGSMGRVGAAGDNAAMESFFSLLQKNVLDRRRWATREELRIAI
MUL_2306|M.ulcerans_Agy99 ------TVGAMGRVQAAGDNAAMASFFSLLRKNVLGRRRWETREHLRIAI
MMAR_3786|M.marinum_M NMRRWGLLGSFGTVGDCFDNAAMESFWARLQVELLNTRKWATTIELAAAM
:*::* * . ***** **:: *: ::*. *:* * .* *:
Mflv_3195|M.gilvum_PYR-GCK VTWIERTYHRRRRQSGLGRLTPIEFEAIMTTPASQAA--
Mvan_5959|M.vanbaalenii_PYR-1 VTWIERTYHRRRRQSGLGRLTPIEFEAIMTTPASQAA--
MSMEG_6888|M.smegmatis_MC2_155 VTWIERTYHRRRRQAGLGRLTPIEFEAIMTTPASQAA--
MAB_2088|M.abscessus_ATCC_1997 VTWIERTYHRRRRQGALGRLTPVEYEAIMTTTASQAA--
MUL_2306|M.ulcerans_Agy99 ATWTETHLRPPPSASRSWPVAPVRFEAIMISRAIQAA--
MMAR_3786|M.marinum_M ADYIDNFYNIERRHSYLGNISPTEFETLWTSTYSIPQLA
. : : . . ::* .:*:: : .