For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSVATEQSTKPTLMLLDGNSLAFRAFYALPAENFKTRGGLTTNAVYGFTAMLINLLRDEAPTHIAAAFDV SRQTFRSERYPEYKANRSSTPDEFHGQIDITKEVLNALGITVLAAPGFEADDIIATLATQAEGEGYRVLV VTGDRDSLQLVSDDVTVLYPRKGVSELTRFTPEAVVEKYGLTPVQYPDFAALRGDPSDNLPGIPGVGEKT ASKWIVEYGSLQGLVDQVDSVRGKVGDSLRANLASVVRNRELTDLVRDVPLAQTPEVLLLQPWDRDQIHR LFDDLEFRVLRDRLFDTLAAVEPEVDQGFDVRGGAVSPGAVAQWLAEHVVDGRRCGLTVVGTHLPHGGDA TALAIAAADGEGGYIDTATLTPDDDAALGEWLADPAKPKALHEAKLAIHDLRGRGWTLNGVTSDTALAAY LVRPGQRSFTLDDLSLRYLRRELRAETPEQQQLSLLDDDDTDVAAAQTAILRARAVIDLADALDAELDRI DSTGLLTDMELPVQHLLAEMETAGIAVDLTMLGGLQRHFADQIRDAAEAAYAVIGKQINLGSPKQLQVVL FDELRMPKTKRTKTGYTTDADALQSLFDKTGHPFLQHLLAHRDVTRLKVTVDGLLNSVADDGRIHTTFNQ TIAATGRLSSTEPNLQNIPIRTDAGRQIRDAFVVGDGYSELMTADYSQIEMRIMAHLSRDEGLIEAFNTG EDLHSFVASRAFGVPIDEVTGELRRRVKAMSYGLAYGLSAYGLSSQLKISTEDAKVQMDQYFSRFGGVRD YLMAVVEQARKDGYTSTVLGRRRHLPELDSSNRQVREAAERAALNAPIQGSAADIIKVAMIEVDKALKEH ALKSRMLLQVHDELLFEVAAGEREQIEALVRDKMGSAYPLDVPLEVSVGFGRSWDSAAH
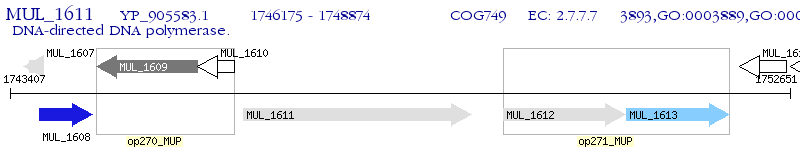
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1611 | polA | - | 100% (899) | DNA polymerase I |
| M. ulcerans Agy99 | MUL_2322 | - | 2e-41 | 37.32% (276) | 5'-3' exonuclease |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1655 | polA | 0.0 | 89.21% (899) | DNA polymerase I |
| M. gilvum PYR-GCK | Mflv_3564 | - | 0.0 | 84.94% (890) | DNA polymerase I |
| M. tuberculosis H37Rv | Rv1629 | polA | 0.0 | 89.21% (899) | DNA polymerase I |
| M. leprae Br4923 | MLBr_01381 | polA | 0.0 | 84.13% (901) | DNA polymerase I |
| M. abscessus ATCC 19977 | MAB_2615c | - | 0.0 | 82.63% (898) | DNA polymerase I |
| M. marinum M | MMAR_2432 | polA | 0.0 | 99.22% (899) | DNA polymerase I PolA |
| M. avium 104 | MAV_3155 | - | 0.0 | 87.18% (897) | DNA polymerase I |
| M. smegmatis MC2 155 | MSMEG_3839 | - | 0.0 | 84.61% (890) | DNA polymerase I |
| M. thermoresistible (build 8) | TH_4211 | - | 0.0 | 84.17% (897) | PUTATIVE DNA polymerase I |
| M. vanbaalenii PYR-1 | Mvan_3361 | - | 0.0 | 83.35% (895) | DNA polymerase I |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3564|M.gilvum_PYR-GCK ----MSPAKTASK-KAATPAADDTPTLMLLDGNSLAFRAFYALPAENFKT
Mvan_3361|M.vanbaalenii_PYR-1 ----MSPAKTASETKAGKPDTSEKPTLMLLDGNSLAFRAFYALPAENFKT
MSMEG_3839|M.smegmatis_MC2_155 ----MSPAKTATK-KTPAKAADDTPTLMLLDGNSLAFRAFYALPAENFKT
TH_4211|M.thermoresistible__bu ----VSPAKTTEQ-PAPAEQTSEQPTLLLLDGNSLAFRAFYALPAENFKT
MAV_3155|M.avium_104 MPATKAATKTATKTAAGAGEEQNRPTLMLLDGNSLAFRAFYALPAENFKT
MUL_1611|M.ulcerans_Agy99 --------------MSVATEQSTKPTLMLLDGNSLAFRAFYALPAENFKT
MMAR_2432|M.marinum_M --------------MSVATEQSTKPTLMLLDGNSLAFRAFYALPAENFKT
Mb1655|M.bovis_AF2122/97 ----------MVTTASAPSEDRAKPTLMLLDGNSLAFRAFYALPAENFKT
Rv1629|M.tuberculosis_H37Rv ----------MVTTASAPSEDRAKPTLMLLDGNSLAFRAFYALPAENFKT
MLBr_01381|M.leprae_Br4923 ---------MSAAMTAEVCEDHTKPMLMLLDGNSLAFRAFYALPTENFKT
MAB_2615c|M.abscessus_ATCC_199 ------------MTAPATKTGTKQPTLLLLDGNSLAFRAFYALPAENFKT
. * *:****************:*****
Mflv_3564|M.gilvum_PYR-GCK QGGLTTNAVYGFTAMLINLIRDEQPSHIAAAFDVSRQTFRKEKYPEYKEG
Mvan_3361|M.vanbaalenii_PYR-1 KSGLTTNAVYGFTAMLINLLRDEQPTHIAAAFDVSRQTFRKDKYPEYKEG
MSMEG_3839|M.smegmatis_MC2_155 QSGLTTNAVYGFTAMLINLLRDEQPTHIAAAFDVSRQTFRKDKYPEYKEG
TH_4211|M.thermoresistible__bu KSGLTTNAVYGFTAMLINLLRDENPTHVAAAFDVSRQTFRLDKYPEYKAG
MAV_3155|M.avium_104 RGGLTTNAVYGFTAMLINLLRDEAPTHIAAAFDVSRQTFRSDRYPEYKAN
MUL_1611|M.ulcerans_Agy99 RGGLTTNAVYGFTAMLINLLRDEAPTHIAAAFDVSRQTFRSERYPEYKAN
MMAR_2432|M.marinum_M RGGLTTNAVYGFTAMLINLLRDEAPTHIAAAFDVSRQTFRSERYPEYKAN
Mb1655|M.bovis_AF2122/97 RGGLTTNAVYGFTAMLINLLRDEAPTHIAAAFDVSRQTFRLQRYPEYKAN
Rv1629|M.tuberculosis_H37Rv RGGLTTNAVYGFTAMLINLLRDEAPTHIAAAFDVSRQTFRLQRYPEYKAN
MLBr_01381|M.leprae_Br4923 RGGLTTNAVYGFTAMLINLLREEAPTHIAAAFDVSRKTFRSECYAGYKAN
MAB_2615c|M.abscessus_ATCC_199 QSGLTTNAVYGFTSMLINLLRDEAPTHVAAAFDVSRKTFRSDRYPEYKAT
:.***********:*****:*:* *:*:********:*** : *. **
Mflv_3564|M.gilvum_PYR-GCK RSATPDEFRGQIDITKEVLGALGITVLAEAGFEADDIIATLATQAEGEGH
Mvan_3361|M.vanbaalenii_PYR-1 RSATPDEFRGQIDITKEVLGALGITVLAEPGFEADDIIATLATQAENEGH
MSMEG_3839|M.smegmatis_MC2_155 RSATPDEFRGQIDITKEVLGALGITVLAEPGFEADDIIATLATQAEQEGY
TH_4211|M.thermoresistible__bu RAATPDEFRGQIEITKEVLAALGITVLAEPGYEADDIIATLATQAEAEGF
MAV_3155|M.avium_104 RSATPDEFHGQIDITKEVLAALGITVLAEPGFEADDLIATLATQAENEGY
MUL_1611|M.ulcerans_Agy99 RSSTPDEFHGQIDITKEVLNALGITVLAAPGFEADDIIATLATQAEGEGY
MMAR_2432|M.marinum_M RSSTPDEFHGQIDITKEVLNALGITVLAAPGFEADDIIATLATQAEGEGY
Mb1655|M.bovis_AF2122/97 RSSTPDEFAGQIDITKEVLGALGITVLSEPGFEADDLIATLATQAENEGY
Rv1629|M.tuberculosis_H37Rv RSSTPDEFAGQIDITKEVLGALGITVLSEPGFEADDLIATLATQAENEGY
MLBr_01381|M.leprae_Br4923 RSSIPAEFHGQIDITKEVLGALGITVFAEAGFEADDLIATLATQAENEGY
MAB_2615c|M.abscessus_ATCC_199 RSATPDEFRGQIDITKEVLQALGITVLAEEGFEADDIIATLATQGQAEGF
*:: * ** ***:****** ******:: *:****:*******.: **.
Mflv_3564|M.gilvum_PYR-GCK RVLVVTGDRDALQLVSDDVTVLYPRKGVSELTRFTPEAVQEKYGLTPAQY
Mvan_3361|M.vanbaalenii_PYR-1 RVLVVTGDRDSLQLVSDDVTVLYPRKGVSELTRFTPEAVQEKYGLTPAQY
MSMEG_3839|M.smegmatis_MC2_155 RVLVVTGDRDSLQLVSDQVTVLYPRKGVSELTRFTPDAVVEKYGLTPQQY
TH_4211|M.thermoresistible__bu RVLIVTGDRDSLQLVTDNVTVLYPRKGVSELTRFTPEAVYEKYGLTPEQY
MAV_3155|M.avium_104 RVLVVTGDRDSLQLVSDNVTVLYPRKGVSELTRFTPEAVVEKYGLTPQQY
MUL_1611|M.ulcerans_Agy99 RVLVVTGDRDSLQLVSDDVTVLYPRKGVSELTRFTPEAVVEKYGLTPVQY
MMAR_2432|M.marinum_M RVLVVTGDRDSLQLVSDDVTVLYPRKGVSELTRFTPEAVVEKYGLTPVQY
Mb1655|M.bovis_AF2122/97 RVLVVTGDRDALQLVSDDVTVLYPRKGVSELTRFTPEAVVEKYGLTPRQY
Rv1629|M.tuberculosis_H37Rv RVLVVTGDRDALQLVSDDVTVLYPRKGVSELTRFTPEAVVEKYGLTPRQY
MLBr_01381|M.leprae_Br4923 RVLVVTGDRDALQLVSNDVTVLYPRKGVSELTRFTPEAVIEKYGVTPAQY
MAB_2615c|M.abscessus_ATCC_199 KVFVVTGDRDSLQLVNEDVTVLYPRKGVSDLTRFTPEAVVEKYGLTPSQY
:*::******:****.::***********:******:** ****:** **
Mflv_3564|M.gilvum_PYR-GCK PDFAALRGDPSDNLPGIPGVGEKTATKWIAEYGSLQGLVDNVDKVKGKVG
Mvan_3361|M.vanbaalenii_PYR-1 PDFAALRGDPSDNLPGIPGVGEKTATKWIAEYGSLQSLVDNVDKVKGKVG
MSMEG_3839|M.smegmatis_MC2_155 PDFAALRGDPSDNLPGIPGVGEKTATKWIVEYGSLQALVDNVDAVKGKVG
TH_4211|M.thermoresistible__bu PDYAALRGDPSDNLPGIPGVGEKTAAKWIREYGSLQGLVDNVDAVRGKVG
MAV_3155|M.avium_104 PDFAALRGDPSDNLPGIPGVGEKTASKWIAEYGSLQALVDNVDSVRGKVG
MUL_1611|M.ulcerans_Agy99 PDFAALRGDPSDNLPGIPGVGEKTASKWIVEYGSLQGLVDQVDSVRGKVG
MMAR_2432|M.marinum_M PDFAALRGDPSDNLPGIPGVGEKTASKWIVEYGSLQGLVDQVDSVRGKVG
Mb1655|M.bovis_AF2122/97 PDFAALRGDPSDNLPGIPGVGEKTAAKWIAEYGSLRSLVDNVDAVRGKVG
Rv1629|M.tuberculosis_H37Rv PDFAALRGDPSDNLPGIPGVGEKTAAKWIAEYGSLRSLVDNVDAVRGKVG
MLBr_01381|M.leprae_Br4923 PDLAALRGDPSDNLPGIPGVGEKTAAKWIVDYGSLQGLVDNVESVRGKVG
MAB_2615c|M.abscessus_ATCC_199 PDFAALRGDPSDNLPGIPGVGEKTASKWILEYGSLQELVDKADTVRGKVG
** **********************:*** :****: ***:.: *:****
Mflv_3564|M.gilvum_PYR-GCK DALRAHLSSVVLNRELTELVKDVPLAQTPDTLRMQPWDRDHIHRLFDDLE
Mvan_3361|M.vanbaalenii_PYR-1 DALRAHLSSVVLNRELTDLVKDVPLAHTPDTLRMQPWDRDQIHRLFDDLE
MSMEG_3839|M.smegmatis_MC2_155 DALRANLSSVILNRELTDLIRDVPLPQTPDTLRMQPWNRDQIHRLFDDLE
TH_4211|M.thermoresistible__bu DALRANLSSVVLNRELTDLVRDVPLAHTPETLRMQPWDRDQIHRLFDDLE
MAV_3155|M.avium_104 DALREHLASVIRNRELTDLIKDVPLAQTPDTLRLQPWDRDQIHRLFDDLE
MUL_1611|M.ulcerans_Agy99 DSLRANLASVVRNRELTDLVRDVPLAQTPEVLLLQPWDRDQIHRLFDDLE
MMAR_2432|M.marinum_M DSLRANLASVVRNRELTDLVRDVPLAQTPEALRLQPWDRDQIHRLFDDLE
Mb1655|M.bovis_AF2122/97 DALRANLASVVRNRELTDLVRDVPLAQTPDTLRLQPWDRDHIHRLFDDLE
Rv1629|M.tuberculosis_H37Rv DALRANLASVVRNRELTDLVRDVPLAQTPDTLRLQPWDRDHIHRLFDDLE
MLBr_01381|M.leprae_Br4923 EALRTHLASVVRNRELTELVKDVPLVQTSDTLRLQPWDRDRIHRLFDNLE
MAB_2615c|M.abscessus_ATCC_199 DSLRANLASVILNRELTDLVRTVPLPYVPAQLALAPWDRDQIHRLFDDLE
::** :*:**: *****:*:: *** .. * : **:**:******:**
Mflv_3564|M.gilvum_PYR-GCK FRVLRDRLFDTLASADP---EVEEGFDVQGEALQAGTLAPWLAENS-DGR
Mvan_3361|M.vanbaalenii_PYR-1 FRVLRDRLFDTLASADP---EVEEGFEVRGEALEAGTLAAWLAEHS-NGQ
MSMEG_3839|M.smegmatis_MC2_155 FRVLRDRLFETLVAVEP---EVEHGFDVRGRALEPGELAAWLSEHS-LGS
TH_4211|M.thermoresistible__bu FRVLRDRLFETLSSADP---EVEHGFDVRGRALEPGELAAWLAEHS-LGN
MAV_3155|M.avium_104 FRVLRDRLFETLAAVEP---EVDEGFDVRGGALEAGELAVWLAEHS-SGK
MUL_1611|M.ulcerans_Agy99 FRVLRDRLFDTLAAVEP---EVDQGFDVRGGAVSPGAVAQWLAEHVVDGR
MMAR_2432|M.marinum_M FRVLRDRLFDTLAAVEP---EVDQGFDVRGGAVSPGAVAQWLAEHVVDGR
Mb1655|M.bovis_AF2122/97 FRVLRDRLFDTLAAAGG--PEVDEGFDVRGGALAPGTVRQWLAEHAGDGR
Rv1629|M.tuberculosis_H37Rv FRVLRDRLFDTLAAAGG--PEVDEGFDVRGGALAPGTVRQWLAEHAGDGR
MLBr_01381|M.leprae_Br4923 FRVLRDRLFEALAAAGERVPEVDEGFDVRGGLLESGTVGRWLAKHADDGR
MAB_2615c|M.abscessus_ATCC_199 FRVLRDRLFETLEAVEP---EVDEGFQLAGQALEPGTVAAWLAEHTGDGR
*********::* :. **:.**:: * : .* : **::: *
Mflv_3564|M.gilvum_PYR-GCK RFGLAVVGNHLAFDSDATALTIVASGGEGRYIDTTGLDPDDEKALASWLA
Mvan_3361|M.vanbaalenii_PYR-1 RFGVAVVGNHLAFDSDATAVALVASDGDGRYIDTTRLDPEDEKALASWLA
MSMEG_3839|M.smegmatis_MC2_155 RFGVAVVGTHKAYDADATALAIVAADGDGRYIDTSTLTPEDEAALASWLA
TH_4211|M.thermoresistible__bu RFGLAVVGTHRTFDSDATALAIVAADGEGRYIDTATLHPDDEAALASWLA
MAV_3155|M.avium_104 RFGLAVAGTHLAYDADCTALAIVSADGEGRYIDTSALDPDDEAALASWLA
MUL_1611|M.ulcerans_Agy99 RCGLTVVGTHLPHGGDATALAIAAADGEGGYIDTATLTPDDDAALGEWLA
MMAR_2432|M.marinum_M RCGLTVVGTHLPHGGDATALAIAAADGEGGYIDTATLTPDDDAALGEWLA
Mb1655|M.bovis_AF2122/97 RAGLTVVGTHLPHGGDATAMAVAAADGEGAYLDTATLTPDDDAALAAWLA
Rv1629|M.tuberculosis_H37Rv RAGLTVVGTHLPHGGDATAMAVAAADGEGAYLDTATLTPDDDAALAAWLA
MLBr_01381|M.leprae_Br4923 RSGLAIVGTHLPHGGDATALAVAAADGNGGYIDTAMLTPDDDDALAAWLA
MAB_2615c|M.abscessus_ATCC_199 RSGLAVVGTYTPYDGDATAVAIASADGDGAYIDTTTVTPDDEAALAEWLA
* *:::.*.: ....*.**:::.::.*:* *:**: : *:*: **. ***
Mflv_3564|M.gilvum_PYR-GCK DPHVPKALHEAKLAIHDLQGRGWTLAGVTSDTALAAYLVRPGQRSFALDD
Mvan_3361|M.vanbaalenii_PYR-1 DPDAPKALHEAKLAMHDLQGRGWTLAGVTSDTALAAYLVRPGQRSFALDD
MSMEG_3839|M.smegmatis_MC2_155 DPGPPKALHEAKLAMHDLAGRGWTLRGVTSDTALAAYLVRPGQRSFTLDD
TH_4211|M.thermoresistible__bu DPGPPKALHEAKQAMHDLAGRGWRLNGVTSDTALAAYLVRPGQRSFALDD
MAV_3155|M.avium_104 DPGPPKALHEAKLAMHDLAGRGWTLAGVTSDTALAAYLVRPGQRSFSLDD
MUL_1611|M.ulcerans_Agy99 DPAKPKALHEAKLAIHDLRGRGWTLNGVTSDTALAAYLVRPGQRSFTLDD
MMAR_2432|M.marinum_M DPAKPKALHEAKLAIHDLRGRGWTLNGVTSDTALAAYLVRPGQRSFTLDD
Mb1655|M.bovis_AF2122/97 DPAKPKALHEAKAAVHDLAGRGWTLEGVTSDTALAAYLVRPGQRSFTLDD
Rv1629|M.tuberculosis_H37Rv DPAKPKALHEAKAAVHDLAGRGWTLEGVTSDTALAAYLVRPGQRSFTLDD
MLBr_01381|M.leprae_Br4923 DPDNPKALHEAKLAMHDLAGRGWTLGGITSDTALAAYLVRPGQRSFTLDD
MAB_2615c|M.abscessus_ATCC_199 DAARPKALHEAKQAIHALAGRGWELGGVTSDTALAAYLVRPGQRSFALDD
*. ******** *:* * **** * *:******************:***
Mflv_3564|M.gilvum_PYR-GCK LSLRYLKRELRADNPEQQQLSLLDDSDGVDDQAVQTLLLRASAVVDLADA
Mvan_3361|M.vanbaalenii_PYR-1 LSLRYLKRELRADNPEQQQLSLLDDSDAVDDQAVQTLLLRAGAVVDLADA
MSMEG_3839|M.smegmatis_MC2_155 LAVRYLHRELRAETPEQQQLSLLDDSDGVDEQAVQTVILRACAVLDLADA
TH_4211|M.thermoresistible__bu LTLRYLRRELRAEEPEQQQLSLLDESDGVDEQAVQTLLLRACAVMDLADA
MAV_3155|M.avium_104 LALRYLRRELRAENPQQQQLSLLDDTDGTDDQAVQTLILRAVAVLDLADA
MUL_1611|M.ulcerans_Agy99 LSLRYLRRELRAETPEQQQLSLLD-DDDTDVAAAQTAILRARAVIDLADA
MMAR_2432|M.marinum_M LSLRYLRRELRAETPEQQQLSLLD-DDDTDVAAAQTAILRARAVIDLADA
Mb1655|M.bovis_AF2122/97 LSLRYLRRELRAETPQQQQLSLLD-DDDTDAETIQTTILRARAVIDLADA
Rv1629|M.tuberculosis_H37Rv LSLRYLRRELRAETPQQQQLSLLD-DDDTDAETIQTTILRARAVIDLADA
MLBr_01381|M.leprae_Br4923 LSLRYLRRELRAETPEQEQFSLLDNVDEVDKQAIQTLILRARAVVDLAAA
MAB_2615c|M.abscessus_ATCC_199 LSLRYLRRELRAEDDGEQQLSLLDEDGAGDAKAAQAQMLRARAVIDLADA
*::***:*****: ::*:**** . * : *: :*** **:*** *
Mflv_3564|M.gilvum_PYR-GCK LDEELARIDSSALLGNMELPVQRVLAELETAGIAVDLEMLSGLQSEFADQ
Mvan_3361|M.vanbaalenii_PYR-1 LDEELARIDSSALLGSMELPVQRVLAQLETVGIAVDLAMLSELQSEFAGQ
MSMEG_3839|M.smegmatis_MC2_155 LDQELARIDSLSLLSRMELPVQRTLAEMEHAGIAVDLGMLEQLQSEFADQ
TH_4211|M.thermoresistible__bu LDEELARIDSLSLLERMELPVQRVLFEMETAGIAVDLDRLGELQSEFAGQ
MAV_3155|M.avium_104 LDEELARINSTSLLSGMELPVQRVLAGMEKAGIAVDLDLLSELQSDFAHQ
MUL_1611|M.ulcerans_Agy99 LDAELDRIDSTGLLTDMELPVQHLLAEMETAGIAVDLTMLGGLQRHFADQ
MMAR_2432|M.marinum_M LDAELDRIDSTGLLADMELPVQHVLAEMETAGIAVDLTMLGGLQSHFADQ
Mb1655|M.bovis_AF2122/97 LDAELARIDSTALLGEMELPVQRVLAKMESAGIAVDLPMLTELQSQFGDQ
Rv1629|M.tuberculosis_H37Rv LDAELARIDSTALLGEMELPVQRVLAKMESAGIAVDLPMLTELQSQFGDQ
MLBr_01381|M.leprae_Br4923 LDAELDLIDSTSLLGEMELPVQQVLADMEKAGIAADLRLLTELQSQFGDQ
MAB_2615c|M.abscessus_ATCC_199 LDVELERIESAGLLTDMELPVQRVLAGLESVGIAVDTDHLSSLQNQFASG
** ** *:* .** ******: * :* .***.* * ** .*.
Mflv_3564|M.gilvum_PYR-GCK IRDAAEAAYAVIGKQINLGSPKQLQVVLFDELEMPKTKRTKTGYTTDADA
Mvan_3361|M.vanbaalenii_PYR-1 IRDAAEAAYAVIGKQINLGSPKQLQVVLFDELEMPKTKRTKTGYTTDADA
MSMEG_3839|M.smegmatis_MC2_155 IRDAAEAAYSVIGKQINLGSPKQLQAVLFDELEMPKTKKTKTGYTTDADA
TH_4211|M.thermoresistible__bu IRDAAEAAYAVIGKQINLGSPKQLQGVLFDELGMPKTKRTKTGYTTDADA
MAV_3155|M.avium_104 IRDAAEAAYAVIGKQINLGSPKQLQAVLFDELQMPKTKRTKTGYTTDADA
MUL_1611|M.ulcerans_Agy99 IRDAAEAAYAVIGKQINLGSPKQLQVVLFDELRMPKTKRTKTGYTTDADA
MMAR_2432|M.marinum_M IRDAAEAAYAVIGKQINLGSPKQLQVVLFDELGMPKTKRTKTGYTTDADA
Mb1655|M.bovis_AF2122/97 IRDAAEAAYGVIGKQINLGSPKQLQVVLFDELGMPKTKRTKTGYTTDADA
Rv1629|M.tuberculosis_H37Rv IRDAAEAAYGVIGKQINLGSPKQLQVVLFDELGMPKTKRTKTGYTTDADA
MLBr_01381|M.leprae_Br4923 IRDAAEAAYAVIGKQINLSSPKQLQVVLFEELGMPKTKRTKTGYTTDADA
MAB_2615c|M.abscessus_ATCC_199 IREAADAAYAVIGKQINLGSPKQLQVVLFDELGMPKTKKTKTGYTTDADA
**:**:***.********.****** ***:** *****:***********
Mflv_3564|M.gilvum_PYR-GCK LQSLFDKTGHPFLQHLLAHRDATRLKVTVDGLLNAVASDGRIHTTFNQTI
Mvan_3361|M.vanbaalenii_PYR-1 LQSLFDKTGHPFLQHLLAHRDATRLKVTVDGLLNSVAADGRIHTTFNQTI
MSMEG_3839|M.smegmatis_MC2_155 LQSLFEKTGHPFLQHLLAHRDATRLKVTVDGLLNSVASDGRIHTTFNQTI
TH_4211|M.thermoresistible__bu LQSLFEKTGHPFLQHLLAHRDATRLKVTVDGLLNSVAADGRIHTTFNQTI
MAV_3155|M.avium_104 LQSLFDKTGHPFLQHLLAHRDATRLKVTVDGLLNSVASDGRIHTTFNQTI
MUL_1611|M.ulcerans_Agy99 LQSLFDKTGHPFLQHLLAHRDVTRLKVTVDGLLNSVADDGRIHTTFNQTI
MMAR_2432|M.marinum_M LQSLFDKTGHPFLQHLLAHRDVTRLKVTVDGLLNSVADDGRIHTTFNQTI
Mb1655|M.bovis_AF2122/97 LQSLFDKTGHPFLQHLLAHRDVTRLKVTVDGLLQAVAADGRIHTTFNQTI
Rv1629|M.tuberculosis_H37Rv LQSLFDKTGHPFLQHLLAHRDVTRLKVTVDGLLQAVAADGRIHTTFNQTI
MLBr_01381|M.leprae_Br4923 LQSLFCKTEHPFLQHLLTHRDVTRLKVTVDGLLNAVAADGRIHTTFNQTI
MAB_2615c|M.abscessus_ATCC_199 LQTLFDKTGHPFLEHLLTHRDVTRLKVTVDGLLKSVAADGRIHTTFNQTI
**:** ** ****:***:***.***********::** ************
Mflv_3564|M.gilvum_PYR-GCK AATGRLSSTEPNLQNIPIRTEAGRRIRDAFVVG---DGYGELMTADYSQI
Mvan_3361|M.vanbaalenii_PYR-1 AATGRLSSTEPNLQNIPIRTEAGRRIRDGFVVG---EGYADLMTADYSQI
MSMEG_3839|M.smegmatis_MC2_155 AATGRLSSTEPNLQNIPIRTEAGRRIRDAFVVG---EGYAELMTADYSQI
TH_4211|M.thermoresistible__bu AATGRLSSTEPNLQNIPIRTEAGRMIRDAFVVG---DGYAELMTADYSQI
MAV_3155|M.avium_104 AATGRLSSTEPNLQNIPIRTEAGRRIRDAFVVG---DGYAELMTADYSQI
MUL_1611|M.ulcerans_Agy99 AATGRLSSTEPNLQNIPIRTDAGRQIRDAFVVG---DGYSELMTADYSQI
MMAR_2432|M.marinum_M AATGRLSSTEPNLQNIPIRTDAGRQIRDAFVVG---DGYSELMTADYSQI
Mb1655|M.bovis_AF2122/97 AATGRLSSTEPNLQNIPIRTDAGRRIRDAFVVG---DGYAELMTADYSQI
Rv1629|M.tuberculosis_H37Rv AATGRLSSTEPNLQNIPIRTDAGRRIRDAFVVG---DGYAELMTADYSQI
MLBr_01381|M.leprae_Br4923 ATTGRLSSTEPNLQNIPIRTNAGRQIRDAFVVGSENNGYTELMTADYSQI
MAB_2615c|M.abscessus_ATCC_199 AATGRLSSTEPNLQNIPVRTEAGREIRDGFVVG---AGYRELMTVDYSQI
*:***************:**:*** ***.**** ** :***.*****
Mflv_3564|M.gilvum_PYR-GCK EMRIMAHLSRDEGLIEAFNTGEDLHSFVASRAFSVPIDEVTAELRRRVKA
Mvan_3361|M.vanbaalenii_PYR-1 EMRIMAHLSKDAGLIEAFNTGEDLHSFVASRAFSVPLDEVTPELRRRVKA
MSMEG_3839|M.smegmatis_MC2_155 EMRIMAHLSRDAGLIEAFNTGEDLHSFVASRAFSVPIDEVTPELRRRVKA
TH_4211|M.thermoresistible__bu EMRIMAHLSGDEGLIEAFRTGEDLHSFVASRAFSVPIEEVTPELRRRVKA
MAV_3155|M.avium_104 EMRIMAHLSQDAGLIEAFNTGEDLHSFVASRAFGVPIEEVTGELRRRVKA
MUL_1611|M.ulcerans_Agy99 EMRIMAHLSRDEGLIEAFNTGEDLHSFVASRAFGVPIDEVTGELRRRVKA
MMAR_2432|M.marinum_M EMRIMAHLSRDEGLIEAFNTGEDLHSFVASRAFGVPIDEVTGELRRRVKA
Mb1655|M.bovis_AF2122/97 EMRIMAHLSGDEGLIEAFNTGEDLHSFVASRAFGVPIDEVTGELRRRVKA
Rv1629|M.tuberculosis_H37Rv EMRIMAHLSGDEGLIEAFNTGEDLHSFVASRAFGVPIDEVTGELRRRVKA
MLBr_01381|M.leprae_Br4923 EMRIMAHLSRDEGLIEAFHTGEDLHSFVASRAFGIPIEDITPELRRRVKA
MAB_2615c|M.abscessus_ATCC_199 EMRIMAHLSADEGLIEAFNTGEDLHSFVAARAFGVPIDEVTAELRRRVKA
********* * ******.**********:***.:*::::* ********
Mflv_3564|M.gilvum_PYR-GCK MSYGLAYGLSAYGLSQQLKISTEEAKEQMEQYFARFGGVRDYLRDVVDQA
Mvan_3361|M.vanbaalenii_PYR-1 MSYGLAYGLSAYGLSQQLKISTEEAKEQMEQYFARFGGVRDYLRDVVDQA
MSMEG_3839|M.smegmatis_MC2_155 MSYGLAYGLSAYGLAQQLKISTEEAKVQMEQYFDRFGGVRDYLRDVVDQA
TH_4211|M.thermoresistible__bu MSYGLAYGLSAYGLAQQLKISTEEAKVQMEQYFARFGKVRDYLQSVVEQA
MAV_3155|M.avium_104 MSYGLAYGLSAYGLSQQLKISTEEAKEQMDAYFARFGGVRDYLHAVVEQA
MUL_1611|M.ulcerans_Agy99 MSYGLAYGLSAYGLSSQLKISTEDAKVQMDQYFSRFGGVRDYLMAVVEQA
MMAR_2432|M.marinum_M MSYGLAYGLSAYGLSSQLKISTEDAKVQMDQYFSRFGGVRDYLMAVVEQA
Mb1655|M.bovis_AF2122/97 MSYGLAYGLSAYGLSQQLKISTEEANEQMDAYFARFGGVRDYLRAVVERA
Rv1629|M.tuberculosis_H37Rv MSYGLAYGLSAYGLSQQLKISTEEANEQMDAYFARFGGVRDYLRAVVERA
MLBr_01381|M.leprae_Br4923 MSYGLAYGLSAYGLATQLKISTEEAKLQMEQYFARFGGVRDYLMDVVEQA
MAB_2615c|M.abscessus_ATCC_199 MSYGLAYGLSAYGLATQLKISNDEAKEQMDQYFSRFGGVRDYLRAVVDQA
**************: *****.::*: **: ** *** ***** **::*
Mflv_3564|M.gilvum_PYR-GCK RKDGYTSTVFGRRRYLPELDSSNRQVREAAERAALNAPIQGSAADIIKVA
Mvan_3361|M.vanbaalenii_PYR-1 RKDGYTSTVFGRRRYLPELDSSNRQVREAAERAALNAPIQGSAADIIKVA
MSMEG_3839|M.smegmatis_MC2_155 RKDGYTSTVLGRRRYLPELDSSNRQVREAAERAALNAPIQGSAADIIKVA
TH_4211|M.thermoresistible__bu RKDGYTSTVLGRRRYLPELDSSNRQVREAAERAALNAPIQGSAADIIKVA
MAV_3155|M.avium_104 RKDGYTSTVLGRRRYLPELDSSNRQVREAAERAALNAPIQGSAADIIKVA
MUL_1611|M.ulcerans_Agy99 RKDGYTSTVLGRRRHLPELDSSNRQVREAAERAALNAPIQGSAADIIKVA
MMAR_2432|M.marinum_M RKDGYTSTVLGRRRYLPELDSSNRQVREAAERAALNAPIQGSAADIIKVA
Mb1655|M.bovis_AF2122/97 RKDGYTSTVLGRRRYLPELDSSNRQVREAAERAALNAPIQGSAADIIKVA
Rv1629|M.tuberculosis_H37Rv RKDGYTSTVLGRRRYLPELDSSNRQVREAAERAALNAPIQGSAADIIKVA
MLBr_01381|M.leprae_Br4923 RKDGYTSTVLGRRRYLPELDSSNRQIREAAERAALNAPIQGSAADIIKVA
MAB_2615c|M.abscessus_ATCC_199 RKDGYTSTVLGRRRYLPDLDSGDRLRREAAERAALNAPIQGSAADIIKVA
*********:****:**:***.:* ************************
Mflv_3564|M.gilvum_PYR-GCK MINVDQAIKDAGLKSRTLLQVHDELLFEVADGERDTLDALVREHMGSAYA
Mvan_3361|M.vanbaalenii_PYR-1 MINVDQAIKEAGLSSRMLLQVHDELLFEVADGERDALDALVREHMGSAYR
MSMEG_3839|M.smegmatis_MC2_155 MINVDQAIKDAGLRSRILLQVHDELLFEVSEGEQGELEQLVREHMGNAYP
TH_4211|M.thermoresistible__bu MINVDEAIKAAGLKSRMLLQVHDELLFEVAPGERDTLEQLVREQMGGAYP
MAV_3155|M.avium_104 MIEVDKAIKEAGLQSRILLQVHDELLFEVAAGEREQLEALARDKMGGAYP
MUL_1611|M.ulcerans_Agy99 MIEVDKALKEHALKSRMLLQVHDELLFEVAAGEREQIEALVRDKMGSAYP
MMAR_2432|M.marinum_M MIEVDKALKEHALKSRMLLQVHDELLFEVAAGEREQIEALVRDKMGSAYP
Mb1655|M.bovis_AF2122/97 MIQVDKALNEAQLASRMLLQVHDELLFEIAPGERERVEALVRDKMGGAYP
Rv1629|M.tuberculosis_H37Rv MIQVDKALNEAQLASRMLLQVHDELLFEIAPGERERVEALVRDKMGGAYP
MLBr_01381|M.leprae_Br4923 MIAVDKSLKQAKLASRMLLQVHDELLFEVAIGEREQIEAMVREQMGSAYP
MAB_2615c|M.abscessus_ATCC_199 MITTDKALRDKGLRSRMLLQVHDELLFEVAEGERESLEALVRKTMGSAYP
** .*:::. * ** ***********:: **: :: :.*. **.**
Mflv_3564|M.gilvum_PYR-GCK LDVPLEVSVGFGRSWDAAAH
Mvan_3361|M.vanbaalenii_PYR-1 LDVPLEVSVGYGRSWDSAAH
MSMEG_3839|M.smegmatis_MC2_155 LDVPLEVSVGYGRSWDAAAH
TH_4211|M.thermoresistible__bu LDVPLEVSVGFGRSWDAAAH
MAV_3155|M.avium_104 LDVPLEVSVGYGRSWDAAAH
MUL_1611|M.ulcerans_Agy99 LDVPLEVSVGFGRSWDSAAH
MMAR_2432|M.marinum_M LDVPLEVSVGFGRSWDSAAH
Mb1655|M.bovis_AF2122/97 LDVPLEVSVGYGRSWDAAAH
Rv1629|M.tuberculosis_H37Rv LDVPLEVSVGYGRSWDAAAH
MLBr_01381|M.leprae_Br4923 LDVPLEVSVGFGRSWGAAAH
MAB_2615c|M.abscessus_ATCC_199 LDVPLEVSVGYGRSWDAAAH
**********:****.:***