For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SVDTVREPLQERSEVIDSRRWPAVAKPPSGPLSAVAAAIAHRLLRRAVARLPLRLVYPDGTSIGAADGPT IVVHQPAALARRIGRNGLIGFGESYMAAEWSSNDLAAALTVFAESVAELIPPSLQWLRPIAPAFRPRWRD PSRDEARRNIAEHYDLSNELFAQFLDDTMTYSSALFDQLPAAWPDLAAAQRRKIDRLLDMAEVRDGSRVL EIGTGWGELCIRAASRGAQVRSITLSVEQQRLARRRVAEASLSHLVQIDLCDYRDVGSEGAAYDSVLSVE MIEAIGYHSWPKYFAALQRLVRPGGKVAIQAITMPHDRMLTSRNTLTWVQKYIFPGGLLPSTQAIADITE RHTALRTIDTASLRPHYAQTLRLWRERFLQRQGELAQLGFDDVFKRMWELYLAYSEAGFRSGYLDVYQWT FEREDGR*
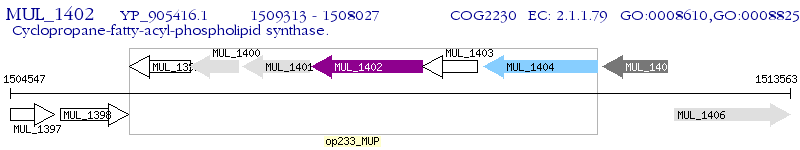
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1402 | ufaA1 | - | 100% (428) | cyclopropane-fatty-acyl-phospholipid synthase UfaA1 |
| M. ulcerans Agy99 | MUL_0730 | mmaA3 | 1e-28 | 28.77% (285) | methoxy mycolic acid synthase 3, MmaA3 |
| M. ulcerans Agy99 | MUL_0729 | mmaA4 | 6e-24 | 28.16% (277) | methoxy mycolic acid synthase 4, MmaA4 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0455c | ufaA1 | 1e-177 | 72.16% (431) | putative cyclopropane-fatty-acyl-phospholipid synthase |
| M. gilvum PYR-GCK | Mflv_1283 | - | 8e-36 | 28.75% (400) | cyclopropane-fatty-acyl-phospholipid synthase |
| M. tuberculosis H37Rv | Rv0447c | ufaA1 | 1e-177 | 72.16% (431) | cyclopropane-fatty-acyl-phospholipid synthase |
| M. leprae Br4923 | MLBr_02334 | - | 2e-34 | 27.61% (402) | hypothetical protein MLBr_02334 |
| M. abscessus ATCC 19977 | MAB_0310c | - | 7e-39 | 29.67% (418) | putative cyclopropane-fatty-acyl-phospholipid synthase |
| M. marinum M | MMAR_0768 | ufaA1 | 0.0 | 98.83% (428) | cyclopropane-fatty-acyl-phospholipid synthase UfaA1 |
| M. avium 104 | MAV_0383 | - | 1e-35 | 29.13% (412) | cyclopropane-fatty-acyl-phospholipid synthase |
| M. smegmatis MC2 155 | MSMEG_6284 | - | 3e-38 | 29.50% (400) | cyclopropane-fatty-acyl-phospholipid synthase |
| M. thermoresistible (build 8) | TH_2292 | - | 4e-37 | 29.37% (395) | POSSIBLE FATTY ACID SYNTHASE |
| M. vanbaalenii PYR-1 | Mvan_5532 | - | 6e-36 | 30.02% (403) | cyclopropane-fatty-acyl-phospholipid synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1283|M.gilvum_PYR-GCK MTTFREH----TDSSASDPDRKLTLAEVLEIFAA-GRRPLKFTAYDGSSC
Mvan_5532|M.vanbaalenii_PYR-1 MTTFRDG----AADTGLHGDRKLTLAEVLEVFAS-GRLPLKFTAYDGSSA
MSMEG_6284|M.smegmatis_MC2_155 MTTFKER----ETSTA---DRKLTLAEILEIFAA-GKEPLKFTAYDGSSA
TH_2292|M.thermoresistible__bu MTTFKEHPGFKEHPGRLATDRKLSLAEIFEIVAD-GDLPIRFTAYDGSAA
MAB_0310c|M.abscessus_ATCC_199 MTTTQAK-------NARGENGKLSLAEILELMAA-GDLPLRFSAYDGSTA
MLBr_02334|M.leprae_Br4923 ------------------------MAEILGILTATGQQPLKFTAYDGSTA
MAV_0383|M.avium_104 MTTTKQP--------HRSRTGKLSMAEILEVFAATGRHPLKFTAYDGSTA
MUL_1402|M.ulcerans_Agy99 MSVDTVREPLQERSEVIDSRRWPAVAKPPS-----GPLSAVAAAIAHRLL
MMAR_0768|M.marinum_M MSVDTVREPLHERSEVIDSRRWPAVAKPPS-----GPLSAVAAAIAHRLL
Mb0455c|M.bovis_AF2122/97 MTVETSQTP----SAAIDSDRWPAVAKVPR-----GPLAAASAAIANRLL
Rv0447c|M.tuberculosis_H37Rv MTVETSQTP----SAAIDSDRWPAVAKVPR-----GPLAAASAAIANRLL
:*: * . :*
Mflv_1283|M.gilvum_PYR-GCK GPEDATLGLDLLTPRGTTYLATAPGDLGLARAYIAGDLRLSGVHPGDPHD
Mvan_5532|M.vanbaalenii_PYR-1 GPDDATLGLDLLTPRGTTYLATAPGDLGLARAYVSGDLQLQGVHPGDPYD
MSMEG_6284|M.smegmatis_MC2_155 GPEDATMGLDLKTPRGTTYLATAPGDLGLARAYVSGDLEPHGVHPGDPYP
TH_2292|M.thermoresistible__bu GPEDARLGIDLLAPRGTTYLATAPGDLGLARAYVSGDLEPRGVHPGDPYE
MAB_0310c|M.abscessus_ATCC_199 GPENAELGLDLLTPRGTTYLATAPGDLGLARAYVSGDIEMQGVHPGDPYE
MLBr_02334|M.leprae_Br4923 GHDDAEIGLDLCSPRGATYLATAPGELGIARAYVAGDLQARGVHPGDPYT
MAV_0383|M.avium_104 GSEDAELGLDLRSPRGATYLATAPGELGLARAYVAGDLQAYGVHPGDPYR
MUL_1402|M.ulcerans_Agy99 RRAVARLPLRLVYPDGTSIGAADG--PTIVVHQPAALARRIGRNGLIGFG
MMAR_0768|M.marinum_M RRAVARLPLRLVYPDGTSIGAADG--PTIVVHQPVALARRIGRNGLIGFG
Mb0455c|M.bovis_AF2122/97 RRTATHLPLRLVYSDGTATGAADPRAPSLFIHRPDALARRIGRHGLIGFG
Rv0447c|M.tuberculosis_H37Rv RRTATHLPLRLVYSDGTATGAADPRAPSLFIHRPDALARRIGRHGLIGFG
: : : * . *:: *: : . . * : .
Mflv_1283|M.gilvum_PYR-GCK LLTALTERLEYRRPPVR-VLANVLRSIGIEHLKPVAPPPQEHLPRWRRIA
Mvan_5532|M.vanbaalenii_PYR-1 LLNALVQKLDFKRPSAR-VLAQVVRSIGIEHLKPIAPPPQEALPRWRRIA
MSMEG_6284|M.smegmatis_MC2_155 LLRALAERMEFKRPPAR-VLANIVRSIGIEHLKPIAPPPQEALPRWRRIM
TH_2292|M.thermoresistible__bu LLKTLDQ-VSFRRPPAR-VLAHIVRSIGFEHLKPIAPPPQEALPRWRRIA
MAB_0310c|M.abscessus_ATCC_199 LLKAMAERLDFKRPPAR-VLANIVRSIGLEHLRPIAPPPQEALPRWRRMV
MLBr_02334|M.leprae_Br4923 LLKTLTDRVQFKRPSLR-VLASVVRSLGVEHLLPIAPPPQETPPRWRRIT
MAV_0383|M.avium_104 LLKTLTDRVQFKRPPAR-VLANVVRSIGFERLLPVAPPPQETRPRWRRIA
MUL_1402|M.ulcerans_Agy99 ESYMAAEWSSNDLAAALTVFAESVAELIPPSLQWLRPIAPAFRPRWR---
MMAR_0768|M.marinum_M ESYMAAEWSSNDLAAALTVFAESVAELIPPSLQWLRPFAPAFRPRWR---
Mb0455c|M.bovis_AF2122/97 ESYMAGEWSSKELTRVLTVLAGSVDELVPRSLHWLRPITPTFRPSWP---
Rv0447c|M.tuberculosis_H37Rv ESYMAGEWSSKELTRVLTVLAGSVDELVPRSLHWLRPITPTFRPSWP---
: . . *:* : .: * : * . * *
Mflv_1283|M.gilvum_PYR-GCK EGLRHSKTRDAEAIQHHYDVSNTFYSWVLGPSMTYTCACYPHPDAT----
Mvan_5532|M.vanbaalenii_PYR-1 EGLRHSKTRDADAIHHHYDVSNTFYEWVLGPSMTYTCACYPHPDAT----
MSMEG_6284|M.smegmatis_MC2_155 EGLRHSKTRDAEAIHHHYDVSNTFYEWVLGPSMTYTCACYPTEDAT----
TH_2292|M.thermoresistible__bu EGLRHSKTRDAEAIHHHYDVSNTFYRYVLGPSMTYTCACYPDADAS----
MAB_0310c|M.abscessus_ATCC_199 EGFRHSKTRDSEAIHHHYDVSNTFYEWVLGPSMTYTCACYPDAEAT----
MLBr_02334|M.leprae_Br4923 DGLLHSKTRDAEAIRHHYDVSNRFYEWVLGPSMTYTCAVYPNAEAT----
MAV_0383|M.avium_104 DGLMHTKARDAEAIHHHYDVSNRFYELVLGPSMTYTCAVYPHPDAT----
MUL_1402|M.ulcerans_Agy99 ---DPSRDEARRNIAEHYDLSNELFAQFLDDTMTYSSALFDQLPA----A
MMAR_0768|M.marinum_M ---DPSRDEARRNIAEHYDLSNELFAQFLDDTMTYSSALFDQLPA----A
Mb0455c|M.bovis_AF2122/97 ---DHSRDQARRNIAVHYDLSNDLFAAFLDETMTYSCAMFTDLLAQPTPA
Rv0447c|M.tuberculosis_H37Rv ---DHSRDQARRNIAVHYDLSNDLFAAFLDETMTYSCAMFTDLLAQPTPA
:: . * ***:** :: .*. :***:.* : *
Mflv_1283|M.gilvum_PYR-GCK ---LEEAQENKYRLVFEKLRLKPGDRLLDVGCGWGGMVRYAARHGVKVLG
Mvan_5532|M.vanbaalenii_PYR-1 ---LEEAQENKYRLVFEKLRLKPGDRLLDVGCGWGGMVRYAARHGVKAIG
MSMEG_6284|M.smegmatis_MC2_155 ---LEEAQDNKYRLVFEKLRLKPGDRLLDVGCGWGGMVRYAARHGVKALG
TH_2292|M.thermoresistible__bu ---LEEAQENKYRLVFEKLRLQPGDRLLDVGCGWGGMVRYAARHGVNAIG
MAB_0310c|M.abscessus_ATCC_199 ---LEEAQENKYRLVFDKLALKAGDRLLDVGCGWGSMVRYAARRGVRATG
MLBr_02334|M.leprae_Br4923 ---LEEAQENKYRLIFEKLRLQPGDRLLDVGCGWGGMVRYAARRGVRVIG
MAV_0383|M.avium_104 ---LEEAQENKYRLIFDKLRLAPGDRLLDVGCGWGGMVRYAARRGVRAIG
MUL_1402|M.ulcerans_Agy99 WPDLAAAQRRKIDRLLDMAEVRDGSRVLEIGTGWGELCIRAASRGAQVRS
MMAR_0768|M.marinum_M WPDLAAAQRRKIDRLLDMAEVRDGSRVLEIGTGWGELCIRAASRGAQVRS
Mb0455c|M.bovis_AF2122/97 WTELAAAQRRKIDRLLDVAGVQQGSHVLEIGTGWGELCIRAAARGAHIRS
Rv0447c|M.tuberculosis_H37Rv WTELAAAQRRKIDRLLDVAGVQQGSHVLEIGTGWGELCIRAAARGAHIRS
* ** .* ::: : *.::*::* *** : ** :*.. .
Mflv_1283|M.gilvum_PYR-GCK VTLSKEQAQWAADAVERDGLGELAEVRHGDYRDVRE--SHFDAVSSLGLT
Mvan_5532|M.vanbaalenii_PYR-1 VTLSREQAQWARAAIERDGLGDLAEVRHSDYRDVRE--SQFDAVSSLGLT
MSMEG_6284|M.smegmatis_MC2_155 VTLSREQATWAQKAIAQEGLTDLAEVRHGDYRDVIE--SGFDAVSSIGLT
TH_2292|M.thermoresistible__bu VTLSREQALWGQKAIAEEGLGQHAEIRHGDYRDVVE--TGFDAVSSLGLT
MAB_0310c|M.abscessus_ATCC_199 VTLSAEQAAWAQKAIADEGLADLAEVRHADYRDTTE--REFDAVSSIGLT
MLBr_02334|M.leprae_Br4923 ATLSAEQAKWARKAIDNEGLAEIAQVRYSDYRDIRE--TEFDAVSSIGLT
MAV_0383|M.avium_104 ATLSAEQAKWAQRRIDDEGLGDLAQVRHSDYRDVAE--TGFDAVSSIGLT
MUL_1402|M.ulcerans_Agy99 ITLSVEQQRLARRRVAEASLSHLVQIDLCDYRDVGSEGAAYDSVLSVEMI
MMAR_0768|M.marinum_M ITLSVEQQRLARRRVAEAGLSHLVQIDLCDYRDVGSEGAAYDSVLSVEMI
Mb0455c|M.bovis_AF2122/97 VTLSVEQQRLARQRVAAAGFGHRVEIDLCDYRDVDG---QYDSVVSVEMI
Rv0447c|M.tuberculosis_H37Rv VTLSVEQQRLARQRVAAAGFGHRVEIDLCDYRDVDG---QYDSVVSVEMI
*** ** . : .: . .:: **** :*:* *: :
Mflv_1283|M.gilvum_PYR-GCK EHIGVANYPSYFRFLKSKLRPGGLLLNHCITRNNNRSHATAG--GFIDRY
Mvan_5532|M.vanbaalenii_PYR-1 EHIGVANYPSYFRFLKSKLRPGGLLLNHCITRHNNRTGPAAG--GFIDRY
MSMEG_6284|M.smegmatis_MC2_155 EHIGVHNYPAYFNFLKSKLRTGGLLLNHCITRPDNRSAPSAG--GFIDRY
TH_2292|M.thermoresistible__bu EHVGVQNYPAYFRFLQSKLRPGGLLLNHCITRPDNRSAATAG--DFIDRY
MAB_0310c|M.abscessus_ATCC_199 EHIGIANYPAYFRFLQSRLKTGGLLLNHCITRPDNRTSAVAG--YFIDRY
MLBr_02334|M.leprae_Br4923 EHIGVKNYPAYFNFLKLKLRTGGLLLNHCITRRDNKSTSFAG--GFTDRY
MAV_0383|M.avium_104 EHIGVKNYPAYFGFLKSKLRTGGLLLNHCITRHDNTSTSFAG--GFTDRY
MUL_1402|M.ulcerans_Agy99 EAIGYHSWPKYFAALQRLVRPGGKVAIQAITMPHDRMLTSRNTLTWVQKY
MMAR_0768|M.marinum_M EAIGYHSWPKYFAALQRLVRPGGKVAIQAITMPHDRMLASRNTLTWVQKY
Mb0455c|M.bovis_AF2122/97 EAVGYRSWPRYFAALEQLVRPGGPVAIQAITMPHHRMLATRHTQTWIQKY
Rv0447c|M.tuberculosis_H37Rv EAVGYRSWPRYFAALEQLVRPGGPVAIQAITMPHHRMLATRHTQTWIQKY
* :* .:* ** *: ::.** : :.** .. . : ::*
Mflv_1283|M.gilvum_PYR-GCK VFPDGELTGSGRIITEMQ-DVGLEVVHEENLRHHYALTLRDWSRNLVAHW
Mvan_5532|M.vanbaalenii_PYR-1 VFPDGELTGSGRIITEIQ-DVGLEVMHEENLRRHYALTLRDWCRNLVQHW
MSMEG_6284|M.smegmatis_MC2_155 VFPDGELTGSGRIITEAQ-DVGLEVIHEENLRNHYAMTLRDWCRNLVEHW
TH_2292|M.thermoresistible__bu VFPDGELTGSGRIITEAQ-NVGLEVLHEENLRHHYALTLRDWCRNLVENW
MAB_0310c|M.abscessus_ATCC_199 VFPDGELTGSGRIITEIQ-NVGLEVLHEENLRHHYALTLKEWCANLVEHW
MLBr_02334|M.leprae_Br4923 VFPDGELTGSGRITTEIQ-EVGLEVLHEENFRHHYAMTLRDWCHNLVEHW
MAV_0383|M.avium_104 VFPDGELTGSGRITCAIQ-DVGFEVLHGENFRHHYAMTLRDWCRNLVEHW
MUL_1402|M.ulcerans_Agy99 IFPGGLLPSTQAIADITERHTALRTIDTASLRPHYAQTLRLWRERFLQRQ
MMAR_0768|M.marinum_M IFPGGLLPSTQAIADITERHTALRTIDTASLRPHYAQTLRLWRERFLQRQ
Mb0455c|M.bovis_AF2122/97 IFPGGLLPSTQAIIDITGQHTGLRIVDAASLRPHYAETLRLWRERFMQRR
Rv0447c|M.tuberculosis_H37Rv IFPGGLLPSTQAIIDITGQHTGLRIVDAASLRPHYAETLRLWRERFMQRR
:**.* *..: * ...:. :. .:* *** **: * .:: .
Mflv_1283|M.gilvum_PYR-GCK D-DAVTEVGLPTAKVWGLYIAASRVGFEQNAIQLHQVLSVKLDERGSDGG
Mvan_5532|M.vanbaalenii_PYR-1 D-EAVAEVGLPTAKVWGLYMAASRVGFEQNSIQLHQVLAVKLDERGGDGG
MSMEG_6284|M.smegmatis_MC2_155 D-EAVEEVGLPTAKVWGLYMAGSRLGFETNVVQLHQVLAVKLDDQGKDGG
TH_2292|M.thermoresistible__bu D-EAVAEVGLPTAKVWGLYMAGSRLGFETNVVQLHQVLAVKLDESGNDGG
MAB_0310c|M.abscessus_ATCC_199 D-EAVAEVGEATAKVWGLYMAGSRLGFERNVVQLHQVLATKLDERGGSE-
MLBr_02334|M.leprae_Br4923 E-DAVAEVGLPIAKVWGLYMAASRVAFEQNNLQLHHILATKVDVWGDNS-
MAV_0383|M.avium_104 D-AAVDEVGLPTAKVWGLYMAASRVAFEQNNLQLHHVLAANVDARGDDD-
MUL_1402|M.ulcerans_Agy99 GELAQLGFDDVFKRMWELYLAYSEAGFRSGYLDVYQWTFEREDGR-----
MMAR_0768|M.marinum_M GELAQLGFDDVFKRMWELYLAYSEAGFRSGYLDVYQWTFEREDGR-----
Mb0455c|M.bovis_AF2122/97 DGLAHLGFDEVFARMWELYLAYSEAGFRSGYLDVYQWTLIREGPP-----
Rv0447c|M.tuberculosis_H37Rv DGLAHLGFDEVFARMWELYLAYSEAGFRSGYLDVYQWTLIREGPP-----
* .. ::* **:* *. .*. . ::::: . .
Mflv_1283|M.gilvum_PYR-GCK LPLRPWWNA
Mvan_5532|M.vanbaalenii_PYR-1 LPLRPWWTA
MSMEG_6284|M.smegmatis_MC2_155 LPLRPWWSA
TH_2292|M.thermoresistible__bu LPLRPWWTP
MAB_0310c|M.abscessus_ATCC_199 LPLRPWWQP
MLBr_02334|M.leprae_Br4923 LPTRPWWTP
MAV_0383|M.avium_104 LPLRPWWSP
MUL_1402|M.ulcerans_Agy99 ---------
MMAR_0768|M.marinum_M ---------
Mb0455c|M.bovis_AF2122/97 ---------
Rv0447c|M.tuberculosis_H37Rv ---------