For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LRVSASLPEFPWDTLAEAKARAGAHPDGIVDLSVGTPVDPVAPLVQEALTAAAFSPGYPATAGTAGLRES AGAALARRYGITELAESAVLPVIGTKEFIAWLPTLLGLGSADAVVVPELAYPTYEVGARLAGARVQWADS LTQLGPQSPALLYLNSPSNPTGRVLGMEHLQKVVEWARGRGVIVASDECYLGLAWDAQPLSVLHPAVCGG DHTGLLAVHSLSKSSSLAGYRAGFVAGDPELIAELLAVRRHAGMMVPTPVQAAMVAALDDDDHEKQQRER YAQRRAVLLPALTSAGFSVDHSEAGLYLWATRGEGCRDTVAWLAERGILVAPGEFYGPAGAQHVRVALTA TKERIAAAVRRLGEP
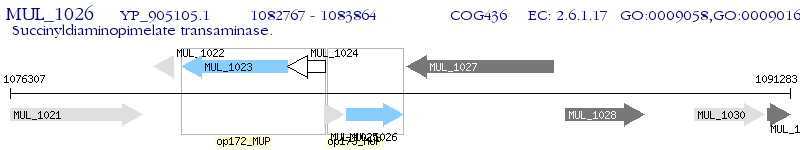
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1026 | - | - | 100% (365) | N-succinyldiaminopimelate aminotransferase |
| M. ulcerans Agy99 | MUL_4130 | aspB | 1e-22 | 26.19% (378) | aspartate aminotransferase |
| M. ulcerans Agy99 | MUL_0297 | - | 1e-21 | 28.02% (389) | aminotransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1211 | - | 1e-178 | 85.28% (360) | N-succinyldiaminopimelate aminotransferase |
| M. gilvum PYR-GCK | Mflv_2171 | - | 1e-162 | 78.12% (361) | N-succinyldiaminopimelate aminotransferase |
| M. tuberculosis H37Rv | Rv1178 | - | 1e-178 | 85.00% (360) | N-succinyldiaminopimelate aminotransferase |
| M. leprae Br4923 | MLBr_01488 | - | 1e-166 | 80.06% (361) | N-succinyldiaminopimelate aminotransferase |
| M. abscessus ATCC 19977 | MAB_1328 | - | 1e-150 | 71.67% (360) | N-succinyldiaminopimelate aminotransferase |
| M. marinum M | MMAR_4273 | - | 0.0 | 99.45% (365) | aminotransferase |
| M. avium 104 | MAV_1317 | - | 1e-171 | 81.94% (360) | N-succinyldiaminopimelate aminotransferase |
| M. smegmatis MC2 155 | MSMEG_5121 | - | 1e-164 | 78.12% (361) | N-succinyldiaminopimelate aminotransferase |
| M. thermoresistible (build 8) | TH_1831 | - | 1e-147 | 79.01% (324) | PROBABLE AMINOTRANSFERASE |
| M. vanbaalenii PYR-1 | Mvan_4528 | - | 1e-163 | 78.59% (355) | N-succinyldiaminopimelate aminotransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2171|M.gilvum_PYR-GCK MRTEPTAELARRRLRAPVSASLPVFPWDTLADVTAAARAHESGIVDLSVG
Mvan_4528|M.vanbaalenii_PYR-1 -----------------------MFPWDTLADATAAARAHPDGIVDLSVG
TH_1831|M.thermoresistible__bu --------------------------------------------------
MSMEG_5121|M.smegmatis_MC2_155 ------------MVRQRRSATLPEFPWDTLADVTALARSHPDGIVDLSVG
MUL_1026|M.ulcerans_Agy99 ----------MR-----VSASLPEFPWDTLAEAKARAGAHPDGIVDLSVG
MMAR_4273|M.marinum_M ----------MR-----VSASLPEFPWDTLAEAKARAGAHPDGIVDLSVG
Mb1211|M.bovis_AF2122/97 -----------------MSASLPVFPWDTLADAKALAGAHPDGIVDLSVG
Rv1178|M.tuberculosis_H37Rv -----------------MSASLPVFPWDTLADAKALAGAHPDGIVDLSVG
MAV_1317|M.avium_104 ------MPHQLRKRRPAVSAALPEFPWDTLAEAKALAGSHPDGIVDLSVG
MLBr_01488|M.leprae_Br4923 ------MSPFGR-----VSATLPVFPWDTLADAKAVAESHPDGIVDLSVG
MAB_1328|M.abscessus_ATCC_1997 -----------------MSASLPEFPWDTIADAKAAAAAHPDGIVDLSVG
Mflv_2171|M.gilvum_PYR-GCK TPVDEVAPVIRDALAAAGSAPGYPTTHGTTALRDAAVAALHRRYGITGLD
Mvan_4528|M.vanbaalenii_PYR-1 TPVDDVAPVIRDALAAASSAPGYPTTHGTPALREAAVSALRRRYGITHLA
TH_1831|M.thermoresistible__bu -----VAPVIRAALAEASAAPGYPTTAGTPALRASAAAALERRYGITGLT
MSMEG_5121|M.smegmatis_MC2_155 TPVDPVAPVIRDALAAGSSSPGYPTTAGTPELREAMVRALDRRYGITGLT
MUL_1026|M.ulcerans_Agy99 TPVDPVAPLVQEALTAAAFSPGYPATAGTAGLRESAGAALARRYGITELA
MMAR_4273|M.marinum_M TPVDPVAPLVQEALTAAAFSPGYPATAGTAGLRESAVAALARRYGITELA
Mb1211|M.bovis_AF2122/97 TPVDPVAPLIQEALAAASAAPGYPATAGTARLRESVVAALARRYGITRLT
Rv1178|M.tuberculosis_H37Rv TPVDPVAPLIQEALAAASAAPGYPATAGTARLRESVVAALARRYGITRLT
MAV_1317|M.avium_104 TPVDPVAPVIQQALAAAGSASGYPATAGTARLRDSAVAALARRFGVVGLT
MLBr_01488|M.leprae_Br4923 TPVDPVAPLIQEALAAASSVAGYPTTAGTARLREAAVAALDRRYGITGLT
MAB_1328|M.abscessus_ATCC_1997 TPVDSVAPVIRDALAAASDLPGYPATAGTPALRQAIRDAVTRRYGVVPLA
***::: **: .. .***:* **. ** : *: **:*:. *
Mflv_2171|M.gilvum_PYR-GCK PAAVLPVIGTKELIAWLPTLLGLGAADTVVVPELAYPTYEVGALLAGAAV
Mvan_4528|M.vanbaalenii_PYR-1 PGSVLPAIGTKELIAWLPTVLGLGAGDTVVVPELAYPTYEVGALLAGAAV
TH_1831|M.thermoresistible__bu PDAVLPAIGTKELIAWLPTLLGLGGDDTVVVPELAYPTYDVGARLAGARV
MSMEG_5121|M.smegmatis_MC2_155 PQTVLPVIGTKELIAWLPTLLGLGSDDVVVVPELAYPTYEVGARLAGAQV
MUL_1026|M.ulcerans_Agy99 ESAVLPVIGTKEFIAWLPTLLGLGSADAVVVPELAYPTYEVGARLAGARV
MMAR_4273|M.marinum_M ESAVLPVIGTKEFIAWLPTLLGLGSADAVVVPELAYPTYEVGARLAGARV
Mb1211|M.bovis_AF2122/97 EAAVLPVIGTKELIAWLPTLLGLGGADLVVVPELAYPTYDVGARLAGTRV
Rv1178|M.tuberculosis_H37Rv EAAVLPVIGTKELIAWLPTLLGLGGADLVVVPELAYPTYDVGARLAGTRV
MAV_1317|M.avium_104 EASVLPVIGSKELIAWLPTLLGLGPADLIVVPELAYPTYDVGARLAGARV
MLBr_01488|M.leprae_Br4923 EAAVLPVIGTKELIAWLPTLLGLRAGDVVVVPELAYPTYNVGARLAGARV
MAB_1328|M.abscessus_ATCC_1997 DNAVLPVIGTKELIAWLPTLLGLGADDLIVIPELAYPTYEVGARLAQVPT
:***.**:**:******:*** * :*:********:*** ** . .
Mflv_2171|M.gilvum_PYR-GCK VRADSLTQLGPAAPPLVYLNSPSNPTGKVLGVDHLRKVVGWARDRGVLVA
Mvan_4528|M.vanbaalenii_PYR-1 ARADSLTQLGPSVPALVYVNSPSNPTGRVLGVDHLRKVVGWARDRGVLVA
TH_1831|M.thermoresistible__bu FRADSLTQLGPQTPALIYLNSPSNPTGRVLGVDHLRKVVGWARDRGVIVA
MSMEG_5121|M.smegmatis_MC2_155 LRADSLTQIGPQVPALIYLNSPSNPTGRVLGRDHLRKVVTWARERGVLVA
MUL_1026|M.ulcerans_Agy99 QWADSLTQLGPQSPALLYLNSPSNPTGRVLGMEHLQKVVEWARGRGVIVA
MMAR_4273|M.marinum_M QRADSLTQLGPQSPALLYLNSPSNPTGRVLGMEHLQKVVEWARGRGVIVA
Mb1211|M.bovis_AF2122/97 LRADALTQLGPQSPALLYLNSPSNPTGRVLGVDHLRKVVEWARGRGVLVV
Rv1178|M.tuberculosis_H37Rv LRADALTQLGPQSPALLYLNSPSNPTGRVLGVDHLRKVVEWARGRGVLVV
MAV_1317|M.avium_104 LRADSLTQVGPQSPALYYLNSPSNPTGRVLGVDHLRKVAAWARDRGCLVA
MLBr_01488|M.leprae_Br4923 LRADSLTQLGLQSPALFYLNSPSNPTGQVLAVNHLRKMVGWARNRGVVVV
MAB_1328|M.abscessus_ATCC_1997 VRADSLTQLGPQRPTLIYLNSPSNPTGAVLPVEHLRKVVEWARERGALVV
**:***:* *.* *:******** ** :**:*:. *** ** :*.
Mflv_2171|M.gilvum_PYR-GCK SDECYLGLGWDATPVSVLHPDVCDGDHTGLLAIHSLSKTSSLAGYRAGFV
Mvan_4528|M.vanbaalenii_PYR-1 SDECYLGLGWDVAPVSVLHPDVCDGDHTGLLAVHSLSKTSSLAGYRAGFV
TH_1831|M.thermoresistible__bu SDECYLGLGWDETPVSVLHPTVCGGDHTNLLAIHSLSKTSSLAGYRAGFV
MSMEG_5121|M.smegmatis_MC2_155 SDECYLGLSWDADPVSVLHPSVCDGDHTGLLAIHSLSKTSSLAGYRAGFV
MUL_1026|M.ulcerans_Agy99 SDECYLGLAWDAQPLSVLHPAVCGGDHTGLLAVHSLSKSSSLAGYRAGFV
MMAR_4273|M.marinum_M SDECYLGLAWDAQPLSVLHPAVCGGDHTGLLAVHSLSKSSSLAGYRAGFV
Mb1211|M.bovis_AF2122/97 SDECYLGLGWDAEPVSVLHPSVCDGDHTGLLAVHSLSKSSSLAGYRAGFV
Rv1178|M.tuberculosis_H37Rv SDECYLGLGWDAEPVSVLHPSVCDGDHTGLLAVHSLSKSSSLAGYRAGFV
MAV_1317|M.avium_104 SDECYLGLGWDAQPLSILHPQVCDGDPTGLLAIHSLSKSSSLAGYRAGFV
MLBr_01488|M.leprae_Br4923 SDECYLGLGWDAEPFSVLHPLVCDGNHTGLLAVHSLSKSSSLAGYRAGFV
MAB_1328|M.abscessus_ATCC_1997 SDECYLGLGWDAEPVSILDPRVCDGETAGLLAVHSLSKTSSLAGYRAGFV
********.** *.*:*.* **.*: :.***:*****:***********
Mflv_2171|M.gilvum_PYR-GCK AGDPAVVAELLAVRKHAGMMVPTPIQAAMVAALDDDAHERTQRARYQRRR
Mvan_4528|M.vanbaalenii_PYR-1 AGDPAVVAELLAVRKHAGMMVPTPIQAAMVAALDDDAHEREQRERYARRR
TH_1831|M.thermoresistible__bu AGDPALVAELLAVRKHAGMMVPTPVQAAMVAALDDDAHERQQRERYQRRR
MSMEG_5121|M.smegmatis_MC2_155 AGDPAVVAELLAVRKHAGMMVPGPIQVAMVAALDDDEHEALQRDRYAQRR
MUL_1026|M.ulcerans_Agy99 AGDPELIAELLAVRRHAGMMVPTPVQAAMVAALDDDDHEKQQRERYAQRR
MMAR_4273|M.marinum_M AGDPELIAELLAVRRHAGMMVPTPVQAAMVAALDDDDHEKQQRERYAQRR
Mb1211|M.bovis_AF2122/97 VGDLEIVAELLAVRKHAGMMVPAPVQAAMVAALDDDAHERQQRERYAQRR
Rv1178|M.tuberculosis_H37Rv VGDLEIVAELLAVRKHAGMMVPAPVQAAMVAALDDDAHERQQRERYAQRR
MAV_1317|M.avium_104 AGDAELIAELLAVRKHAGMMVPTPVQAAMVAALDDDAHERAQRQRYERRR
MLBr_01488|M.leprae_Br4923 AGDPDLVAELLAVRKHAGMMVPTPVQAAMVAALGDDAHEREQRDRYARRR
MAB_1328|M.abscessus_ATCC_1997 LGDGSVVGELLAVRKHAGMMVPGPVQAAMTAALSEDEHQREQRARYEHRR
** ::.******:******* *:*.**.***.:* *: ** ** :**
Mflv_2171|M.gilvum_PYR-GCK DALLPALVSAGFTVDHSEAGLYLWATRDEPCRDTLAWLAQRGILVAPGEF
Mvan_4528|M.vanbaalenii_PYR-1 DALLPALLAAGFTVDHSEAGLYLWATRGEPCRDTVTWLAQKGILVAPGEF
TH_1831|M.thermoresistible__bu RELLPALQQAGFTVDHSEAGLYLWATRGEPCRATVDWLARRGILVAPGEF
MSMEG_5121|M.smegmatis_MC2_155 NLLLPAFREAGFTVDHSEAGLYLWATRGEACRDTLRWLAERGILVAPGEF
MUL_1026|M.ulcerans_Agy99 AVLLPALTSAGFSVDHSEAGLYLWATRGEGCRDTVAWLAERGILVAPGEF
MMAR_4273|M.marinum_M AVLLPALTSAGFSVDHSEAGLYLWATRGEGCRDTVAWLAERGILVAPGEF
Mb1211|M.bovis_AF2122/97 AALLPALGSAGFAVDYSDAGLYLWATRGEPCRDSVAWLAQRGILVAPGDF
Rv1178|M.tuberculosis_H37Rv AALLPALGSAGFAVDYSDAGLYLWATRGEPCRDSAAWLAQRGILVAPGDF
MAV_1317|M.avium_104 AALLPALRSAGFAVDHSEAGLYLWVTRGEPCRDTITWLARRGILAAPGEF
MLBr_01488|M.leprae_Br4923 DVLLPAVVSAGFTVDHSEAGLYLWASRGEPCRDSVEWFAQRGILVALGEF
MAB_1328|M.abscessus_ATCC_1997 DRLSKALTDNGFRIDHSEAGLYLWATRGDSCDDALAYLARRGILVAPGRF
* *. ** :*:*:******.:*.: * : ::*.:***.* * *
Mflv_2171|M.gilvum_PYR-GCK YGPRGTRHVRVALTATDERIDAAVERLGS---
Mvan_4528|M.vanbaalenii_PYR-1 YGPRGARHVRVALTATDERIAAAVARLADSAR
TH_1831|M.thermoresistible__bu YGPRGAEHVRIALTAPDERITAAAARLAQAG-
MSMEG_5121|M.smegmatis_MC2_155 YGPAGDRYVRVALTATDERIQQAAERLAG---
MUL_1026|M.ulcerans_Agy99 YGPAGAQHVRVALTATKERIAAAVRRLGEP--
MMAR_4273|M.marinum_M YGPAGAQHVRVALTATKERIAAAVRRLGEP--
Mb1211|M.bovis_AF2122/97 YGPGGAQHVRVALTATDERVAAAVGRLTC---
Rv1178|M.tuberculosis_H37Rv YGPGGAQHVRVALTATDERVAAAVGRLTC---
MAV_1317|M.avium_104 YGPRGAQHVRVALTADDERIAAAVRRLA----
MLBr_01488|M.leprae_Br4923 YGPGGCQHVRVALTASDERIAAVVARLS----
MAB_1328|M.abscessus_ATCC_1997 YGPRGREHVRVALTATDERIDAAAHRLTAS--
*** * .:**:**** .**: .. **