For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTGPIAVRTHGSSSTRVALALGSGGARGYAHIGVIEALRERGYEIAGIAGSSMGAVVGGLQAAGRLGEFS DWAKSLNQRTILRLLDPSITAAGVLRAEKILDAVRDILGPVTIEQLPVPYTAVATDILAGKSVWFQRGPL DEAIHASIAIPGIIAPHEVDGRLLADGGILDPLPMAPIAGVNADLTIAVSLNGSESGPSRESEPGVTTEW LNRMVRTTSSIFDATAARSLLDRPTRAVLNRFGAGSPDQDSWPEDAQIEAPVPERDDGGYTPGLPKMGSF EVMNRTIDIAQSALARHTLAAYPADLLIEVPRSTCRSLEFHRASEVIAVGRALANRALDALESADQPATP AIEG
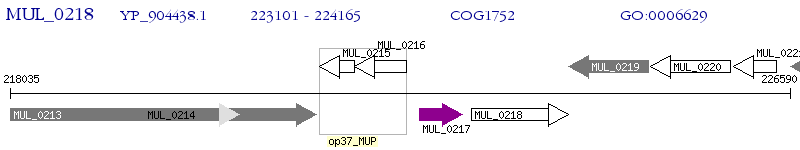
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0218 | - | - | 100% (354) | hypothetical protein MUL_0218 |
| M. ulcerans Agy99 | MUL_3590 | - | 3e-27 | 31.85% (248) | transmembrane transport protein |
| M. ulcerans Agy99 | MUL_1736 | - | 1e-15 | 31.02% (187) | alpha-beta hydrolase superfamily esterase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1092c | - | 1e-158 | 79.17% (360) | hypothetical protein Mb1092c |
| M. gilvum PYR-GCK | Mflv_2030 | - | 1e-117 | 66.67% (330) | patatin |
| M. tuberculosis H37Rv | Rv1063c | - | 1e-158 | 79.17% (360) | hypothetical protein Rv1063c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4751 | - | 6e-94 | 54.89% (348) | hypothetical protein MAB_4751 |
| M. marinum M | MMAR_4403 | - | 0.0 | 99.72% (354) | hypothetical protein MMAR_4403 |
| M. avium 104 | MAV_1190 | - | 1e-138 | 73.62% (345) | patatin |
| M. smegmatis MC2 155 | MSMEG_5284 | - | 1e-122 | 67.93% (343) | patatin |
| M. thermoresistible (build 8) | TH_0679 | - | 1e-113 | 63.91% (327) | CONSERVED HYPOTHETICAL PROTEIN |
| M. vanbaalenii PYR-1 | Mvan_4687 | - | 1e-122 | 68.88% (331) | patatin |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2030|M.gilvum_PYR-GCK --------------MRVALALGSGGARGYAHIGVLNELRERGHEVVGIAG
Mvan_4687|M.vanbaalenii_PYR-1 --------------MRVALALGSGGARGYAHIGVLNELTERGHEVVGIAG
MUL_0218|M.ulcerans_Agy99 MTGPIAVRTHGSSSTRVALALGSGGARGYAHIGVIEALRERGYEIAGIAG
MMAR_4403|M.marinum_M MTGPIAVRTHGSSSTRVALALGSGGARGYAHIGVIEALRERGYEIAGIAG
Mb1092c|M.bovis_AF2122/97 MPAPAALRVRGSSSPRVALALGSGGARGYAHIGVIQALRERGYDIVGIAG
Rv1063c|M.tuberculosis_H37Rv MPAPAALRVRGSSSPRVALALGSGGARGYAHIGVIQALRERGYDIVGIAG
MAV_1190|M.avium_104 ----------------MALALGSGGARGYAHIGVIEALQARGYDIVGVAG
MSMEG_5284|M.smegmatis_MC2_155 --------MTHNESMRVALALGSGSARGYAHIGVINELRARGYEIVGISG
TH_0679|M.thermoresistible__bu ----------------VALALGSGGARGYAHIGVIEELRARGHEVVGVSG
MAB_4751|M.abscessus_ATCC_1997 ---MTGEASTGVPGTKIALALGAGGARGYAHIGVIQELHERGFEISGISG
:*****:*.*********:: * **.:: *::*
Mflv_2030|M.gilvum_PYR-GCK SSMGALVGGLEAAGKTDEFTLWATSLTQRAVLRLLDPSISSPGVLRAEKI
Mvan_4687|M.vanbaalenii_PYR-1 SSMGALVGGLQAAGKLDEYTAWASSLTQRAVLRLLDPSITSAGVLRAEKI
MUL_0218|M.ulcerans_Agy99 SSMGAVVGGLQAAGRLGEFSDWAKSLNQRTILRLLDPSITAAGVLRAEKI
MMAR_4403|M.marinum_M SSMGAVVGGLQAAGRLGEFSDWAKSLNQRTILRLLDPSITAAGVLRAEKI
Mb1092c|M.bovis_AF2122/97 SSMGAVVGGVHAAGRLDEFAHWAKSLTQRTILRLLDPSISAAGILRAEKI
Rv1063c|M.tuberculosis_H37Rv SSMGAVVGGVHAAGRLDEFAHWAKSLTQRTILRLLDPSISAAGILRAEKI
MAV_1190|M.avium_104 SSMGALVGGLQAAGRLDEFAEWAKSLTQRTILRLLDPSISAAGVMRAGKI
MSMEG_5284|M.smegmatis_MC2_155 SSMGALVGGLYAAGKLDEFAEWACTLTQRAVLRLLDPSLSAAGILRAEKI
TH_0679|M.thermoresistible__bu SSMGALVGGVMAAGKLDDYAAWARTLTQRSVLRLLDPSLTAPGVLRAEKI
MAB_4751|M.abscessus_ATCC_1997 SSMGALVGGLEAAGALDEFALWATSLTQRAVLRLLDPTWTSPGFFRAEKI
*****:***: *** .::: ** :*.**::******: ::.*.:** **
Mflv_2030|M.gilvum_PYR-GCK LESVRDILGEVTIEDLPIPYTSVATDLITGKSVWMQRGRVDDAIRASIAI
Mvan_4687|M.vanbaalenii_PYR-1 LDAVRAIVGDVTIEELPIPYTAVATDLITGKSVWMQRGPLDDAIRASIAI
MUL_0218|M.ulcerans_Agy99 LDAVRDILGPVTIEQLPVPYTAVATDILAGKSVWFQRGPLDEAIHASIAI
MMAR_4403|M.marinum_M LDAVRDILGPVTIEQLPVPYTAVATDILAGKSVWFQRGPLDEAIRASIAI
Mb1092c|M.bovis_AF2122/97 LDAVRDIVGPVAIEQLPIPYTAVATDLLAGKSVWFQRGPLDAAIRASIAI
Rv1063c|M.tuberculosis_H37Rv LDAVRDIVGPVAIEQLPIPYTAVATDLLAGKSVWFQRGPLDAAIRASIAI
MAV_1190|M.avium_104 LDAVRDILGPVAIEDLRIPYTAVATDLLAGKSVWFQRGPLDEAIRASIAI
MSMEG_5284|M.smegmatis_MC2_155 LDAVRDILGDVTIEELPIPYTAVATDLITGKAVWLQRGRVDTAIRASIAI
TH_0679|M.thermoresistible__bu LDAVREILGEATIETLPIPFTAVATDLIAGKSVWLQRGPVDGAIRASIAI
MAB_4751|M.abscessus_ATCC_1997 LDVVRELLGDVAIEDLPIPFTAVATDLIAGKSVWLQNGSLASAIRASIAI
*: ** ::* .:** * :*:*:****:::**:**:*.* : **:*****
Mflv_2030|M.gilvum_PYR-GCK PGLITPHVLGGRLLADGGILDPLPMAPIAAAPADITIAVSLSGEDPA---
Mvan_4687|M.vanbaalenii_PYR-1 PGVITPYVLDGRLLADGGILDPLPMAPIAAVAADMTIAVSISGDDPE---
MUL_0218|M.ulcerans_Agy99 PGIIAPHEVDGRLLADGGILDPLPMAPIAGVNADLTIAVSLNGSESG---
MMAR_4403|M.marinum_M PGIIAPHEVDGRLLADGGILDPLPMAPIAGVNADLTIAVSLNGSESG---
Mb1092c|M.bovis_AF2122/97 PGVIAPHEVDGRLLADGGILDPLPMAPIAGVNADLTIAVSLNGSEAG---
Rv1063c|M.tuberculosis_H37Rv PGVIAPHEVDGRLLADGGILDPLPMAPIAGVNADLTIAVSLNGSEAG---
MAV_1190|M.avium_104 PGVIAPHTLDGRLLADGGILDPLPMAPLSAVNADLTIAVNLSGSEAI---
MSMEG_5284|M.smegmatis_MC2_155 PGVIAPHVHDGRLLGDGGILDPLPMAPIASVNADLTIAVSVSGSETPPET
TH_0679|M.thermoresistible__bu PGVIAPHVLDGRLLADGGILDPVPMAPIAAVNADVTIAVSLAGSEAG---
MAB_4751|M.abscessus_ATCC_1997 PGMISPHVLNGRVLADGGVLDLPAVAPLAGVHAELRVAVTLSGGDER---
**:*:*: .**:*.***:** .:**::.. *:: :**.: * :
Mflv_2030|M.gilvum_PYR-GCK ------VDRRMQDLRQPTTDWLSRMWRSSSSLLDTKAARSLLETPTGRAV
Mvan_4687|M.vanbaalenii_PYR-1 ------ASGRGRESRAPSTDWLNRMWRSSSSLLDTKAARSLLDTPTGRAV
MUL_0218|M.ulcerans_Agy99 --------PSRESEPGVTTEWLNRMVRTTSSIFDATAARSLLDRPT-RAV
MMAR_4403|M.marinum_M --------PSRESEPGVTTEWLNRMVRTTSSIFDATAARSLLDRPT-RAV
Mb1092c|M.bovis_AF2122/97 --------PARDAEPNVTAEWLNRMVRSTSALFDVSAARSLLDRPTARAV
Rv1063c|M.tuberculosis_H37Rv --------PARDAEPNVTAEWLNRMVRSTSALFDVSAARSLLDRPTARAV
MAV_1190|M.avium_104 --------TRREAEPAATAEWLTRMMRSTSALFDTAAARSLLDRPTARAV
MSMEG_5284|M.smegmatis_MC2_155 ASEAHSEPAE-GGGTKPTTEWLNRMMRSTSAVLDTAAVRSVLDRPTSRAV
TH_0679|M.thermoresistible__bu ----GAEPADRGAGARPTAEWVN---RLTAALLDSAAARTVRDR------
MAB_4751|M.abscessus_ATCC_1997 RAPAPPEPEPEPREPRVTTEWLNRLLRSTSALFSTDDPE-----------
:::*:. * :::::. .
Mflv_2030|M.gilvum_PYR-GCK LGRFVAADDEPIDAPPEVVKHD--------SADATGVPE----LPRLGSF
Mvan_4687|M.vanbaalenii_PYR-1 LGRFAAGDDEPLPEPDDKVTEE--------PGNAG--------IPKLGAF
MUL_0218|M.ulcerans_Agy99 LNRFGAGSPDQDSWPEDAQIEA----PVPERDDGGYTPG----LPKMGSF
MMAR_4403|M.marinum_M LNRFGAGSPDQDSWPEDAQIEA----PVPERDDGGYTPG----LPKMGSF
Mb1092c|M.bovis_AF2122/97 LSRFGAAAAESDSWSQAPEIEQRPAGPPADREEAADTPG----LPKMGSF
Rv1063c|M.tuberculosis_H37Rv LSRFGAAAAESDSWSQAPEIEQRPAGPPADREEAADTPG----LPKMGSF
MAV_1190|M.avium_104 LSRLGGPSGDADAWPDNPEEPDQP-VLADDDDEAVELPGAATDVPKLGSF
MSMEG_5284|M.smegmatis_MC2_155 LSRFGVSVTESD-DGTDASEDA------TVPESSAEPAG----VPRLGSF
TH_0679|M.thermoresistible__bu ---FGSGAAPADPDPVDQDREL------EEAAENSRSIT----VPRLGSF
MAB_4751|M.abscessus_ATCC_1997 ----VAGQ-------PDP-------------------------AAKLTSF
.:: :*
Mflv_2030|M.gilvum_PYR-GCK EVMNRVIDIAQSALARHTLAAYPPDLLIEVPRTVCRSLDFHRAHEVIEIG
Mvan_4687|M.vanbaalenii_PYR-1 EVMNRVIDIAQAALARHTLAAYPPDLLIEVPRTVCRSLDFHRATEVIEIG
MUL_0218|M.ulcerans_Agy99 EVMNRTIDIAQSALARHTLAAYPADLLIEVPRSTCRSLEFHRASEVIAVG
MMAR_4403|M.marinum_M EVMNRTIDIAQSALARHTLAAYPADLLIEVPRSTCRSLEFHRASEVIAVG
Mb1092c|M.bovis_AF2122/97 EVMNRTIDIAQSALARHTLAGYPADLLIEVPRSTCRSLEFHRAVEVIAVG
Rv1063c|M.tuberculosis_H37Rv EVMNRTIDIAQSALARHTLAGYPADLLIEVPRSTCRSLEFHRAVEVIAVG
MAV_1190|M.avium_104 EVMNRTIDIAQAALARHTLAVYPPDLLIEVPRSICRGLEFHRAVEVIDAG
MSMEG_5284|M.smegmatis_MC2_155 EVMYRTIDIAQAALARHTLAAYPPDLLIEVPRTVCRSLEFNRAAEVIETG
TH_0679|M.thermoresistible__bu EVMNRTIDIAQAALARHTLAAYPPDVVIEIPRSVCRSLDFHRAAEVIDAG
MAB_4751|M.abscessus_ATCC_1997 EVMNRTIEVMQSQLARMQLAAHPPDVLIEVPRSVSRSLDFHRATEVIEVG
*** *.*:: *: *** ** :*.*::**:**: .*.*:*:** *** *
Mflv_2030|M.gilvum_PYR-GCK QELASAALEALDPEPSEA-------
Mvan_4687|M.vanbaalenii_PYR-1 QELASAALDTLEQKVPETASDT---
MUL_0218|M.ulcerans_Agy99 RALANRALDALES-ADQPATPAIEG
MMAR_4403|M.marinum_M RALANRALDALES-ADQPATPAIEG
Mb1092c|M.bovis_AF2122/97 RALATQALEAFEIDDDESAAATIEG
Rv1063c|M.tuberculosis_H37Rv RALATQALEAFEIDDDESAAATIEG
MAV_1190|M.avium_104 RALADQALDALEDGDDDR--PAVER
MSMEG_5284|M.smegmatis_MC2_155 RRLAAEALDS---------------
TH_0679|M.thermoresistible__bu RELAAAALDDAGLT-----------
MAB_4751|M.abscessus_ATCC_1997 RRCAAEALDAYAHTN----------
: * **: