For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSEHAGEIRVAIVGVGNCASSLVQGVQYYRNADENTTVPGLMHVKFGPYHVRDVNFVAAFDVDAKKVGFD LSEAIFASENNTIKIADVPPTDVIVQRGPTLDGIGKYYADTIEVSDAEPVDVVKVLKEAEVDVLVSYLPV GSEEADKFYAQCAIDAGVAFVNALPVFIASDPVWAKKFEDAGVPIVGDDIKSQVGATITHRVMAKLFEDR GVTLDRTYQLNVGGNMDFLNMLERSRLESKKVSKTQAVTSNLSGALAGKVEDKNVHIGPSDHVAWLDDRK WAYVRLEGRAFGDVPLNLEYKLEVWDSPNSAGVIIDAVRAAKIAKDRGIGGPIEAASAYLMKSPPKQLAD DVARAELETFIEG
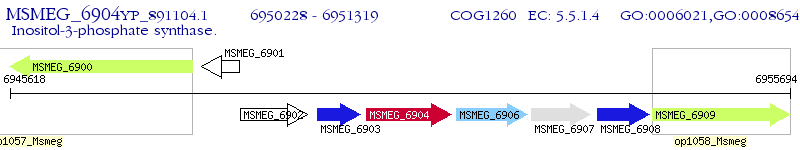
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6904 | - | - | 100% (363) | myo-inositol-1-phosphate synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0047c | ino1 | 0.0 | 87.53% (361) | myo-inositol-1-phosphate synthase INO1 (inositol 1-phosphate |
| M. gilvum PYR-GCK | Mflv_0867 | - | 0.0 | 93.04% (359) | myo-inositol-1-phosphate synthase |
| M. tuberculosis H37Rv | Rv0046c | ino1 | 1e-180 | 87.26% (361) | myo-inositol-1-phosphate synthase INO1 (inositol 1-phosphate |
| M. leprae Br4923 | MLBr_02692 | - | 1e-173 | 81.57% (369) | hypothetical protein MLBr_02692 |
| M. abscessus ATCC 19977 | MAB_4907 | - | 0.0 | 92.01% (363) | myo-inositol-1-phosphate synthase |
| M. marinum M | MMAR_0065 | ino1 | 0.0 | 90.99% (355) | myo-inositol-1-phosphate synthase Ino1 |
| M. avium 104 | MAV_0067 | - | 0.0 | 89.64% (357) | myo-inositol-1-phosphate synthase |
| M. thermoresistible (build 8) | TH_1311 | - | 0.0 | 88.71% (363) | MYO-INOSITOL-1-PHOSPHATE SYNTHASE INO1 (Inositol 1-phosphate |
| M. ulcerans Agy99 | MUL_0064 | ino1 | 0.0 | 91.27% (355) | myo-inositol-1-phosphate synthase Ino1 |
| M. vanbaalenii PYR-1 | Mvan_6040 | - | 0.0 | 94.15% (359) | myo-inositol-1-phosphate synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0867|M.gilvum_PYR-GCK MT--------ASN-DVRVAIVGVGNCASSLVQGVQYYKDADENASVPGLM
Mvan_6040|M.vanbaalenii_PYR-1 MT--------ASN-DVRVAIVGVGNCASSLVQGVQYYKDADENATVPGLM
MSMEG_6904|M.smegmatis_MC2_155 MS--------EHAGEIRVAIVGVGNCASSLVQGVQYYRNADENTTVPGLM
MAB_4907|M.abscessus_ATCC_1997 MS--------NTSSEVRVAIVGVGNCASSLVQGVQYYQDADENSTVPGLM
MMAR_0065|M.marinum_M MSEKKPLGAPGAQPEVRVAIVGVGNCASSLVQGVEYYQNADDNSMVPGLM
MUL_0064|M.ulcerans_Agy99 MSEKKPLGAPGAQPEVRVAIVGVGNCASSLVQGVEYYQNADDNSMVPGLM
Mb0047c|M.bovis_AF2122/97 MSEHQSLPAPEASTEVRVAIVGVGNCASSLVQGVEYYYNADDTSTVPGLM
Rv0046c|M.tuberculosis_H37Rv MSEHQSLPAPEASTEVRVAIVGVGNCASSLVQGVEYYYNADDTSTVPGLM
MLBr_02692|M.leprae_Br4923 MSEHNPAGAPGASTEVRVAIVGVGNCASSLVQGIEYYQNADDTSTVPGLM
MAV_0067|M.avium_104 MSDHQPPQAEDAQSEVRVAIVGVGNCASSLVQGVQYYFDADENSTVPGLM
TH_1311|M.thermoresistible__bu MSEQK---------DIRVAIVGVGNCAASLVQGVEYYKDADENATVPGLM
*: ::***********:*****::** :**:.: *****
Mflv_0867|M.gilvum_PYR-GCK HVKLGAYHVRDVKFVAAFDVDAKKVGFDLSEAIFASENNTIKIADVPPTD
Mvan_6040|M.vanbaalenii_PYR-1 HVKLGPYHVRDVKFVAAFDVDAKKVGFDLSEAIFASENNTIKIADVPPTD
MSMEG_6904|M.smegmatis_MC2_155 HVKFGPYHVRDVNFVAAFDVDAKKVGFDLSEAIFASENNTIKIADVPPTD
MAB_4907|M.abscessus_ATCC_1997 HVKFGPYHVRDVKFVAAFDVDAKKVGFDLSEAISASENNTIKIADVPPTD
MMAR_0065|M.marinum_M HVRFGPYHVRDVKFVAAFDVDAKKVGFDLSDAIFASENNTIKIADVAPTN
MUL_0064|M.ulcerans_Agy99 HVRFGPYHVRDVKFVAAFDVDAKKVGFDLSDAIFASENNTIKIADVAPTN
Mb0047c|M.bovis_AF2122/97 HVRFGPYHVRDVKFVAAFDVDAKKVGFDLSDAIFASENNTIKIADVAPTN
Rv0046c|M.tuberculosis_H37Rv HVRFGPYHVRDVKFVAAFDVDAKKVGFDLSDAIFASENNTIKIADVAPTN
MLBr_02692|M.leprae_Br4923 HVRFGPYHVRDVKFVAAFDVDAKKVGVDLSDAIFTSENNTIKIADVPPTN
MAV_0067|M.avium_104 HVKFGQYHVRDVKFVAAFDVDAKKVGFDLSEAIFASENNTIKIADVPPTN
TH_1311|M.thermoresistible__bu HVRFGPYHVRDVNFVAAFDVDAKKVGFDLSEAIVASENNTIKIADVPPTD
**::* ******:*************.***:** :***********.**:
Mflv_0867|M.gilvum_PYR-GCK VVVQRGPTLDGIGKYYADTIEISDADAVDVVKVLKDAKVDVMVSYLPVGS
Mvan_6040|M.vanbaalenii_PYR-1 VVVQRGPTLDGIGKYYADTIEISDAEAVDVVKVLKDAKVDVLVSYLPVGS
MSMEG_6904|M.smegmatis_MC2_155 VIVQRGPTLDGIGKYYADTIEVSDAEPVDVVKVLKEAEVDVLVSYLPVGS
MAB_4907|M.abscessus_ATCC_1997 VIVQRGPTLDGIGKYYADTIEVSDSEPVDVVKVLKDNNVDVLVSYLPVGS
MMAR_0065|M.marinum_M AVVQRGPTLDGIGKYYADTIELSDDEPVDVVKVLKDNKVDVLVSYLPVGS
MUL_0064|M.ulcerans_Agy99 VVVQRGPTLDGIGKYYADTIELSDDEPVDVVKVLKDNKVDVLVSYLPVGS
Mb0047c|M.bovis_AF2122/97 VIVQRGPTLDGIGKYYADTIELSDAEPVDVVQALKEAKVDVLVSYLPVGS
Rv0046c|M.tuberculosis_H37Rv VIVQRGPTLDGIGKYYADTIELSDAEPVDVVQALKEAKVDVLVSYLPVGS
MLBr_02692|M.leprae_Br4923 VVVQRGPTLDGIGKYYTDTIELSDVPPVDVVQALTEAKTDVLVSYLPVGS
MAV_0067|M.avium_104 VTVQRGPTLDGIGKYYADTIEVSDAEPVDVVQVLKEAKVDVLVSYLPVGS
TH_1311|M.thermoresistible__bu VVVQRGPTLDGIGKYYADTIEVSDAEPVDVVKALKEAEVDVLVSYLPVGS
. **************:****:** .****:.*.: :.**:********
Mflv_0867|M.gilvum_PYR-GCK EEADKFYAQCAIDAGVAFVNALPVFIASDPVWAKKFEDAGVPIVGDDIKS
Mvan_6040|M.vanbaalenii_PYR-1 EEADKFYAQCAIDAGVAFVNALPVFIASDPVWAKKFEDAGVPIVGDDIKS
MSMEG_6904|M.smegmatis_MC2_155 EEADKFYAQCAIDAGVAFVNALPVFIASDPVWAKKFEDAGVPIVGDDIKS
MAB_4907|M.abscessus_ATCC_1997 EEADKFYAQCAIDAKVAFVNALPVFIASDPVWAKKFEDAGVPIVGDDIKS
MMAR_0065|M.marinum_M EEADKFYAQCAIDAGVAFVNALPVFIASDPAWAKKFKDAGVPIIGDDIKS
MUL_0064|M.ulcerans_Agy99 EEADKFYAQCAIDAGVAFVNALPVFIASDPAWAKKFKDAGVPIIGDDIKS
Mb0047c|M.bovis_AF2122/97 EEADKFYAQCAIDAGVAFVNALPVFIASDPVWAKKFTDAGVPIVGDDIKS
Rv0046c|M.tuberculosis_H37Rv EEADKFYAQCAIDAGVAFVNALPVFIASDPVWAKKFTDARVPIVGDDIKS
MLBr_02692|M.leprae_Br4923 EEADKFYAQCAINARVAFVNALPVFIASDPVWAKKFADARVPIVGDDIKS
MAV_0067|M.avium_104 EEADKFYAQCAIDAGVAFVNALPVFIASDPVWAKKFTDAGVPIVGDDIKS
TH_1311|M.thermoresistible__bu EEADKFYAQCCIDAGVAFVNALPVFIASDPVWAKKFADAGVPIVGDDIKS
**********.*:* ***************.***** ** ***:******
Mflv_0867|M.gilvum_PYR-GCK QIGATITHRVMAKLFEDRGVTLDRTYQLNVGGNMDFLNMLERTRLESKKV
Mvan_6040|M.vanbaalenii_PYR-1 QIGATITHRVMAKLFEDRGVTLDRTYQLNVGGNMDFLNMLERSRLESKKV
MSMEG_6904|M.smegmatis_MC2_155 QVGATITHRVMAKLFEDRGVTLDRTYQLNVGGNMDFLNMLERSRLESKKV
MAB_4907|M.abscessus_ATCC_1997 QVGATITHRVMAKLFEDRGVTLDRTYQLNVGGNMDFKNMLERERLESKKV
MMAR_0065|M.marinum_M QVGATITHRVMAKLFEDRGVTLDRTYQLNVGGNMDFKNMLERERLESKKV
MUL_0064|M.ulcerans_Agy99 QVGATITHRVMAKLFEDRGVTLDRTYQLNVGGNMDFKNMLERERLESKKV
Mb0047c|M.bovis_AF2122/97 QVGATITHRVLAKLFEDRGVQLDRTMQLNVGGNMDFLNMLERERLESKKI
Rv0046c|M.tuberculosis_H37Rv QVGATITHRVLAKLFEDRGVQLDRTMQLNVGGNMDFLNMLERERLESKKI
MLBr_02692|M.leprae_Br4923 QVGATITHRVMAKLFEDRGVQLDRTMQLNVGGNMDFLNMLERERVQSKKI
MAV_0067|M.avium_104 QIGATITHRVMAKLFEDRGVQLDRTMQLNVGGNMDFLNMLERERLESKKI
TH_1311|M.thermoresistible__bu QIGATITHRVMAKLFEDRGVQLDRTMQLNVGGNMDFLNMLERERLESKKI
*:********:********* **** ********** ***** *::***:
Mflv_0867|M.gilvum_PYR-GCK SKTQAVTSNLSGSLAGKVEDKNVHIGPSDHVAWLDDRKWAYVRLEGRAFG
Mvan_6040|M.vanbaalenii_PYR-1 SKTQAVTSNLSGSLAGKIEDKNVHIGPSDHVAWLDDRKWAYVRLEGRAFG
MSMEG_6904|M.smegmatis_MC2_155 SKTQAVTSNLSGALAGKVEDKNVHIGPSDHVAWLDDRKWAYVRLEGRAFG
MAB_4907|M.abscessus_ATCC_1997 SKTQAVTSNLNGSLADKVYDKNVHIGPSDYVAWLDDRKWAYVRLEGRAFG
MMAR_0065|M.marinum_M SKTQAVTSNLTGSLAGKVEDKNVHIGPSDHVAWLDDRKWAYVRLEGRAFG
MUL_0064|M.ulcerans_Agy99 SKTQAVTSNLTGSLAGKVEDKNVHIGPSDHVAWLDDRKWAYVRLEGRAFG
Mb0047c|M.bovis_AF2122/97 SKTQAVTSNLKREFK----TKDVHIGPSDHVGWLDDRKWAYVRLEGRAFG
Rv0046c|M.tuberculosis_H37Rv SKTQAVTSNLKREFK----TKDVHIGPSDHVGWLDDRKWAYVRLEGRAFG
MLBr_02692|M.leprae_Br4923 SKTQAVTSNVHREFN----TKDIHIGPSDHVGWLDDRKWAYVRLEGRGFG
MAV_0067|M.avium_104 SKTQAVTSNLQREFK----TKDVHIGPSDHVSWLDDRKWAYVRLEGRAFG
TH_1311|M.thermoresistible__bu SKTQAVTSNLQKEFK----TKDVHIGPSDHVGWLDDRKWAYVRLEGRAFG
*********: : *::******:*.***************.**
Mflv_0867|M.gilvum_PYR-GCK DVPLNLEYKLEVWDSPNSAGVIIDAVRAAKIAKDRGIGGPIEAASAYLMK
Mvan_6040|M.vanbaalenii_PYR-1 DVPLNLEYKLEVWDSPNSAGVIIDAVRAAKIAKDRGIGGPIEAASAYLMK
MSMEG_6904|M.smegmatis_MC2_155 DVPLNLEYKLEVWDSPNSAGVIIDAVRAAKIAKDRGIGGPIEAASAYLMK
MAB_4907|M.abscessus_ATCC_1997 DVPLNLEYKLEVWDSPNSAGIIIDAVRAAKIALDRGLGGPILPASAYLMK
MMAR_0065|M.marinum_M DVPLNLEYKLEVWDSPNSAGIIIDAVRAAKIAKDRGIGGPVIPASAYLMK
MUL_0064|M.ulcerans_Agy99 DVPLNLEYKLEVWDSPNSAGIIIDAVRAAKIAKDRGIGGPVIPASAYLMK
Mb0047c|M.bovis_AF2122/97 DVPLNLEYKLEVWDSPNSAGVIIDAVRAAKIAKDRGIGGPVIPASAYLMK
Rv0046c|M.tuberculosis_H37Rv DVPLNLEYKLEVWDSPNSAGVIIDAVRAAKIAKDRGIGGPVIPASAYLMK
MLBr_02692|M.leprae_Br4923 DVPLNLEYKLEVWDSPNSAGVIIDAVRAAKIAKDRGIGGPVIPASSYLMK
MAV_0067|M.avium_104 DVPLNLEYKLEVWDSPNSAGVIIDAVRAAKIAKDRGIGGPVIPASAYLMK
TH_1311|M.thermoresistible__bu DVPLNLEYKLEVWDSPNSAGVIIDAIRAAKIAKDRGIGGPIESASAYLMK
********************:****:****** ***:***: .**:****
Mflv_0867|M.gilvum_PYR-GCK SPPKQLADDIARVQLEKFIEG--
Mvan_6040|M.vanbaalenii_PYR-1 SPPKQLADDIARVQLEKFIEG--
MSMEG_6904|M.smegmatis_MC2_155 SPPKQLADDVARAELETFIEG--
MAB_4907|M.abscessus_ATCC_1997 SPPKQLADDVARAELEQFIAG--
MMAR_0065|M.marinum_M SPPKQLADDVARTQLEEFIIEG-
MUL_0064|M.ulcerans_Agy99 SPPKQLADDVARTQLEEFIIEG-
Mb0047c|M.bovis_AF2122/97 SPPEQLPDDIARAQLEEFIIG--
Rv0046c|M.tuberculosis_H37Rv SPPEQLPDDIARAQLEEFIIG--
MLBr_02692|M.leprae_Br4923 SPPQQLPDEIARAQLEEFIIDTK
MAV_0067|M.avium_104 SPPQQLPDDVARTQLEEFIIGG-
TH_1311|M.thermoresistible__bu SPPTQMPDDIARAKLEEFIKG--
*** *:.*::**.:** **