For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PDARALGAAQLWIGTSWKMNKGLAESRGYARELAEYVAAKPPAGVQPFIIPSFTALTTVRDALGDDSPVL LGVQNAHWEDHGAWTGEVSVAQAKDAGAQIVEIGHSERREHFGETVETTRLKVAAALHHGLVPLLCIGES AENKQAGESSRFILEQAAGALEGLTDEHLARVLIAYEPIWAIGENGRPATVEELRQPFDDLAREYGCRTM GLLYGGSVNTDNAEDLLGIDHVTGLFIGRAAWQLPGYVRILEMAAAHPKAKA*
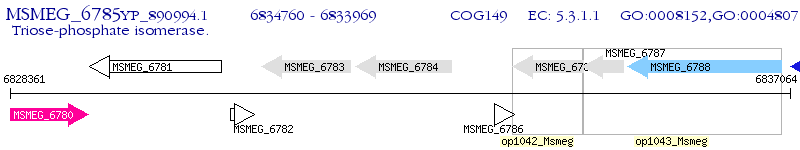
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6785 | tpiA | - | 100% (263) | triosephosphate isomerase |
| M. smegmatis MC2 155 | MSMEG_3086 | tpiA | 2e-34 | 35.04% (254) | triosephosphate isomerase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1473 | tpiA | 5e-35 | 35.83% (254) | triosephosphate isomerase |
| M. gilvum PYR-GCK | Mflv_3708 | tpiA | 1e-36 | 35.83% (254) | triosephosphate isomerase |
| M. tuberculosis H37Rv | Rv1438 | tpiA | 5e-35 | 35.83% (254) | triosephosphate isomerase |
| M. leprae Br4923 | MLBr_00572 | tpiA | 6e-33 | 33.07% (254) | triosephosphate isomerase |
| M. abscessus ATCC 19977 | MAB_2777c | - | 1e-34 | 33.86% (254) | triosephosphate isomerase (TpiA) |
| M. marinum M | MMAR_2241 | tpiA | 4e-33 | 34.65% (254) | triosephosphate isomerase Tpi |
| M. avium 104 | MAV_3339 | tpiA | 3e-34 | 34.65% (254) | triosephosphate isomerase |
| M. thermoresistible (build 8) | TH_0428 | tpi | 1e-34 | 34.65% (254) | PROBABLE TRIOSEPHOSPHATE ISOMERASE TPI (TIM) |
| M. ulcerans Agy99 | MUL_1830 | tpiA | 3e-33 | 34.65% (254) | triosephosphate isomerase |
| M. vanbaalenii PYR-1 | Mvan_2705 | tpiA | 1e-33 | 33.86% (254) | triosephosphate isomerase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3708|M.gilvum_PYR-GCK -------MTRKPLIAGNWKMNLNHFEAIALVQKIAFSLPDKYFDRVDVTV
Mvan_2705|M.vanbaalenii_PYR-1 -------MSRKPLIAGNWKMNLNHFEAIALVQKIAFSLPDKYFDKVDVTV
TH_0428|M.thermoresistible__bu -------VSRKPLIAGNWKMNLNHFEAIALVQKIAFALPAKYFDKVDVAV
MAB_2777c|M.abscessus_ATCC_199 -------MSRKPLIAGNWKMNLNHFEAIALVQKIAFSLPDKYFDKVDVTV
Mb1473|M.bovis_AF2122/97 -------MSRKPLIAGNWKMNLNHYEAIALVQKIAFSLPDKYYDRVDVAV
Rv1438|M.tuberculosis_H37Rv -------MSRKPLIAGNWKMNLNHYEAIALVQKIAFSLPDKYYDRVDVAV
MMAR_2241|M.marinum_M -------MSRKPLIAGNWKMNLNHFEAIALVQKIAFSLPDKYYDKVDVTV
MUL_1830|M.ulcerans_Agy99 -------MSRKPLIAGNWKMNLNHFEAIALVQKIAFSLPDKYYDKVDVTV
MAV_3339|M.avium_104 -------MSRKPLIAGNWKMNLNHFEAIALVQKIAFALPDKYYDKVDVTV
MLBr_00572|M.leprae_Br4923 -------MSRKSLIAGNWKMNLNHFEAIALVQKIAFSLPDKYYDKVDVTV
MSMEG_6785|M.smegmatis_MC2_155 MPDARALGAAQLWIGTSWKMNKGLAESRGYARELAEYVAAKPPAGVQPFI
: : *. .**** . *: . .:::* :. * *: :
Mflv_3708|M.gilvum_PYR-GCK IPPFTDLRSVQTLVDGDKLRLTYGAQDVSQHDSGAYTGEISGAFLAKLGC
Mvan_2705|M.vanbaalenii_PYR-1 IPPFTDLRSVQTLVDGDKLRLTYGAQDVSQHDSGAYTGEISGAFLAKLGC
TH_0428|M.thermoresistible__bu LPPFTDLRSVQTLVDGDDLRLTYGAQDLSQHDSGAYTGEISGAFLAKLGC
MAB_2777c|M.abscessus_ATCC_199 IPPFTDIRSVQTLVDGDKLRLTYGAQDLSVYDSGAYTGEVSGAFLAKLGV
Mb1473|M.bovis_AF2122/97 IPPFTDLRSVQTLVDGDKLRLTYGAQDLSPHDSGAYTGDVSGAFLAKLGC
Rv1438|M.tuberculosis_H37Rv IPPFTDLRSVQTLVDGDKLRLTYGAQDLSPHDSGAYTGDVSGAFLAKLGC
MMAR_2241|M.marinum_M IPPFTDLRSVQTLVDGDKLRLTYGGQDLSQHDSGAYTGDISGAFLAKLGC
MUL_1830|M.ulcerans_Agy99 IPPFTDLRSVQTLVDGDKLRLTYGGQDLSQHDSGAYTGDISGAFLAKLGC
MAV_3339|M.avium_104 LPPFTDLRSVQTLVDGDKLRLSYGAQDLSQHDSGAYTGDISGAFLAKLGC
MLBr_00572|M.leprae_Br4923 LPPFTDLRSVQTLVDGDKLRLTYGAQDLSQHDLGAYTGDISGAFLAKLGC
MSMEG_6785|M.smegmatis_MC2_155 IPSFTALTTVRDALGDDSP-VLLGVQNAHWEDHGAWTGEVSVAQAKDAGA
:*.** : :*: :..*. : * *: * **:**::* * . *
Mflv_3708|M.gilvum_PYR-GCK TFAVVGHSERRTYHGETDELVAAKATAALKHGLTPIVCIGEQLEVREAGN
Mvan_2705|M.vanbaalenii_PYR-1 TFAVVGHSERRTYHNEDDALVAAKAAAAFKHGLTPIICIGEHLDVREAGN
TH_0428|M.thermoresistible__bu TYVIVGHSERRTYHHEDDALVAAKARAAFRHDLIPIVCIGEPLEVREAGN
MAB_2777c|M.abscessus_ATCC_199 TYVVVGHSERRQYHGEDDALVAAKAAAALKHGLTPIVCIGEALDIREAGD
Mb1473|M.bovis_AF2122/97 SYVVVGHSERRTYHNEDDALVAAKAATALKHGLTPIVCIGEHLDVREAGN
Rv1438|M.tuberculosis_H37Rv SYVVVGHSERRTYHNEDDALVAAKAATALKHGLTPIVCIGEHLDVREAGN
MMAR_2241|M.marinum_M SFVVVGHSERRTYHNEDDALVAAKAAAALKHDLTPIVCIGEHLDVREAGN
MUL_1830|M.ulcerans_Agy99 SFVVVGHSERRTYHNEDDALVAAKAAAALKHDLTPIVCIGEHLDVREAGN
MAV_3339|M.avium_104 TFVVVGHSERRTYHNEDDALVAAKAAAALKHELTPIICIGEHLEVREAGN
MLBr_00572|M.leprae_Br4923 SFVLVGHSERRTYHDEGDALVAAKTAAALKNSLTPIVCIGEYLEIREVGE
MSMEG_6785|M.smegmatis_MC2_155 QIVEIGHSERREHFGETVETTRLKVAAALHHGLVPLLCIGESAENKQAGE
. :****** :. * . *. :*::: * *::**** : ::.*:
Mflv_3708|M.gilvum_PYR-GCK HVEFNVNSLRGSLAGLSAEQIAQTVIAYEPVWAIG-TGRVASAADAQEVC
Mvan_2705|M.vanbaalenii_PYR-1 HVEYNVNQLRGSLAGLSAEQIGQAVIAYEPVWAIG-TGRVASAADAQEVC
TH_0428|M.thermoresistible__bu HVAHNLEMLRGSLAGLSAEQIGRAVIAYEPVWAIG-TGRVASAADAQEVC
MAB_2777c|M.abscessus_ATCC_199 HVQYNVNSLRGSLAGLSAEQVGKVVIAYEPVWAIG-TGRVASAADAQEVC
Mb1473|M.bovis_AF2122/97 HVAHNIEQLRGSLAGLLAEQIGSVVIAYEPVWAIG-TGRVASAADAQEVC
Rv1438|M.tuberculosis_H37Rv HVAHNIEQLRGSLAGLLAEQIGSVVIAYEPVWAIG-TGRVASAADAQEVC
MMAR_2241|M.marinum_M HVAHNVEQLRGSLSGLSAEQISKVVIAYEPVWAIG-TGRVAGAADAQEVC
MUL_1830|M.ulcerans_Agy99 HVAHNVEQLRGSLSGLSAEQISKVVIAYEPVWAIG-TGRVAGAADAQEVC
MAV_3339|M.avium_104 HVIHCEEQLRGSLAGLSAEQIGKVVIAYEPVWAIG-TGRVASASDAQEVC
MLBr_00572|M.leprae_Br4923 HVSHCQNQLRGSLAGLSPEQIGNVVIVYEPVWAIG-TGRVASAADAQEVC
MSMEG_6785|M.smegmatis_MC2_155 SSRFILEQAAGALEGLTDEHLARVLIAYEPIWAIGENGRPATVEELRQPF
. : *:* ** *::. .:*.***:**** .** * . : ::
Mflv_3708|M.gilvum_PYR-GCK KAIRDELGNLASADVAAGVRVLYGGSVNAKNVGEIVGQGDVDGALVGGAS
Mvan_2705|M.vanbaalenii_PYR-1 KAIRDELGNLASPELAAGVRVLYGGSVNAKNVGEIVGQADVDGALVGGAS
TH_0428|M.thermoresistible__bu KAIRDELGNLTSPEIAAGVRVLYGGSVNAKNVGEIVAQEDVDGALVGGAS
MAB_2777c|M.abscessus_ATCC_199 AAIRAELAQIANADVAGSVRVLYGGSANAKNVGEIVAQEDVDGALVGGAS
Mb1473|M.bovis_AF2122/97 AAIRKELASLASPRIADTVRVLYGGSVNAKNVGDIVAQDDVDGGLVGGAS
Rv1438|M.tuberculosis_H37Rv AAIRKELASLASPRIADTVRVLYGGSVNAKNVGDIVAQDDVDGGLVGGAS
MMAR_2241|M.marinum_M AAIRSELGSLASPQIADAVRVLYGGSVNAKNIGELIAQADVDGGLVGGAS
MUL_1830|M.ulcerans_Agy99 AAIRSELGSLASPQIADAVRVLYGGSVNAKNIGELIAQADVDGGLVGGAS
MAV_3339|M.avium_104 AAIRKELASLASAQIADSVRVLYGGSVNAKNVGELIAQDDIDGGLVGGAS
MLBr_00572|M.leprae_Br4923 EAIRKELGALASPQVAETVRVLYGGSLNAKNIGDIVAQQDVDGGLVGGAS
MSMEG_6785|M.smegmatis_MC2_155 DDLAREYG-------CRTMGLLYGGSVNTDNAEDLLGIDHVTGLFIGRAA
: * . . : :***** *:.* :::. .: * ::* *:
Mflv_3708|M.gilvum_PYR-GCK LDGEQFATLSAIAAGGPLP--
Mvan_2705|M.vanbaalenii_PYR-1 LDGEQFSMLSAIAAGGPLP--
TH_0428|M.thermoresistible__bu LDGEQFATLSAIAAGGPLP--
MAB_2777c|M.abscessus_ATCC_199 LDGEQFAQMSAIAAGGPLL--
Mb1473|M.bovis_AF2122/97 LDGEHFATLAAIAAGGPLP--
Rv1438|M.tuberculosis_H37Rv LDGEHFATLAAIAAGGPLP--
MMAR_2241|M.marinum_M LDGEQFATLAAIAAGGPLP--
MUL_1830|M.ulcerans_Agy99 LDGEQFATLAAIAAGGPLP--
MAV_3339|M.avium_104 LDGEQFATLAAIAAGGPLP--
MLBr_00572|M.leprae_Br4923 LDGAQFATLAVIAAGGPLP--
MSMEG_6785|M.smegmatis_MC2_155 WQLPGYVRILEMAAAHPKAKA
: : : :**. *