For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SITVDGAPIRRVGINPVAKLAAALVIALGLVLSVDWVSAATALVLEIVLMLVLKVPVRTLLTRGALVLLA AALTGVTILLYGEQSGTVHWHFLLITVSDGSISLALATFLRVLAIALPSVILFIDTDPTELADGLGQVLR LPARFVLGALAGLRMVGLLGQDWRYLGYARRARGVADQDRLRRFAAQGFALLVSAVRRGSMLATAMEARG FGAHPRRTWARPSRFGMRESALILAAFVIAAAAISVSVATGHWNFIGHR*
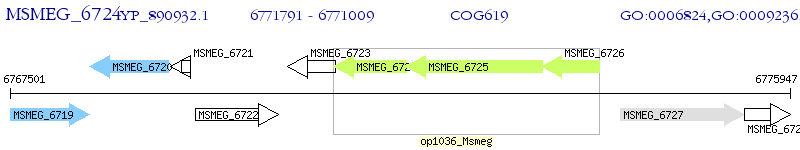
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6724 | - | - | 100% (260) | ABC-type cobalt transport system |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0972 | - | 1e-98 | 73.88% (245) | cobalt transport protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1714 | - | 3e-05 | 23.51% (251) | hypothetical protein MAB_1714 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3956 | - | 3e-06 | 27.10% (262) | cobalt transport protein, putative |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_1714|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_3956|M.avium_104 MAGLTVPTTGTCLLDGRPAAEQVGAVALQFQAARLQLMRSRVDLEVASAA
MSMEG_6724|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0972|M.gilvum_PYR-GCK --------------------------------------------------
MAB_1714|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_3956|M.avium_104 GFSPDDHDRVSAALATVGLDAGLAERRIDQLSGGQMRRVVLAGLLARSPR
MSMEG_6724|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0972|M.gilvum_PYR-GCK --------------------------------------------------
MAB_1714|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_3956|M.avium_104 VLILDEPLAGLDAASQRGLVELLAERRRETGLTVVVISHDFAGLEQLCPR
MSMEG_6724|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0972|M.gilvum_PYR-GCK --------------------------------------------------
MAB_1714|M.abscessus_ATCC_1997 --------------------------MTKTRAPRRPVVLLRPVPGTSTIH
MAV_3956|M.avium_104 ILHLRDGSLDANPVAAQPVPVPVAPPTKRPAARRRPVVLLRPVPGSSPIH
MSMEG_6724|M.smegmatis_MC2_155 ---------------------------MSITVDGAPIRRVGINP------
Mflv_0972|M.gilvum_PYR-GCK --------------------------------------MIRVNP------
: *
MAB_1714|M.abscessus_ATCC_1997 ELWAGTKLIAVFLISVLLTFFPTWTAIGAVAALVILTAWLAHIPRGAVPT
MAV_3956|M.avium_104 ELWAGTKLLVVFAMSLLLTVFPGWVAIGLATALAAAGLRLAHIPRGVLPS
MSMEG_6724|M.smegmatis_MC2_155 ----VAKLAAALVIALGLVLSVDWVSAATALVLEIVLMLVLKVPVRTLLT
Mflv_0972|M.gilvum_PYR-GCK ----VAKLLAALVIAGVLVLSVDWVSALTALVLEIPLFLAMRIPLRAFLI
:** ..: :: *.. *.: . .* ::* ..
MAB_1714|M.abscessus_ATCC_1997 IPIWMVALVIGGSLTVFFSG---GPPFWHIGGLTIGFHGVLAVARTTAVA
MAV_3956|M.avium_104 VPRWLWIFLGVVGVTAALAG---GAPTIHLGTASLGLGGLLDFLRATALT
MSMEG_6724|M.smegmatis_MC2_155 RGALVLLAAALTGVTILLYGEQSGTVHWHFLLITVSD-GSISLALATFLR
Mflv_0972|M.gilvum_PYR-GCK RGAFITLAAALTALTNLLYGEPGGAVYWHFLLVTVSD-GSVELAAAMFLR
: .:* : * *. *: ::. * : . : :
MAB_1714|M.abscessus_ATCC_1997 AVLLGTGAMVSWTTNVSEIAPAVAVLGRPLKVFGIPVDERAATLALALRS
MAV_3956|M.avium_104 VVLLGLGALVSWTTNVAQIAPAVAALGRPLRVLRIPVDDWSVALALALRT
MSMEG_6724|M.smegmatis_MC2_155 VLAIALPSVILFID--TDPTELADGLGQVLRLP----ARFVLGALAGLRM
Mflv_0972|M.gilvum_PYR-GCK VLAIALPSVCLFID--VSPTELADGLGQIMRLP----ARFVVGALAGMRL
.: :. :: : . : . **: ::: .:*
MAB_1714|M.abscessus_ATCC_1997 FPMLSDEFRVLIAARRLRPPQVREGRRGRRGLRWIAEAVDMLTAAITAAL
MAV_3956|M.avium_104 FPMLIDEFRVLYAARRLRPRRPAQTRWAR-LRRPATDLIDVVVAVITVTL
MSMEG_6724|M.smegmatis_MC2_155 VGLLGQDWRYLGYARRARG-VADQDRLRR--------FAAQGFALLVSAV
Mflv_0972|M.gilvum_PYR-GCK VGLLRQDWEYLGYARRARG-VADHARLRR--------FIGQAFALLVFAL
. :* :::. * *** * . * * * :. ::
MAB_1714|M.abscessus_ATCC_1997 RRASEMGDAITARGG---TGQISAHPARPGLNDAIALLLVIAVGTAVVLF
MAV_3956|M.avium_104 RRADEMGDAITARGG---TGQISAAPSRPKRNDWIALSIASAVCAAAVAA
MSMEG_6724|M.smegmatis_MC2_155 RRGSMLATAMEARGFGAHPRRTWARPSRFGMRESALILAAFVIAAAAISV
Mflv_0972|M.gilvum_PYR-GCK RRGSKLATAMEARGFGAYPTRTWARPSTFGVPEMVLIAGAVVIAAAAVAV
**.. :. *: *** . : * *: : : . .: :*.:
MAB_1714|M.abscessus_ATCC_1997 EVL----------
MAV_3956|M.avium_104 ELALLAGH-----
MSMEG_6724|M.smegmatis_MC2_155 SVATGHWNFIGHR
Mflv_0972|M.gilvum_PYR-GCK STATGHWDFIVGR
.