For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MIRVNPVAKLLAALVIAGVLVLSVDWVSALTALVLEIPLFLAMRIPLRAFLIRGAFITLAAALTALTNLL YGEPGGAVYWHFLLVTVSDGSVELAAAMFLRVLAIALPSVCLFIDVSPTELADGLGQIMRLPARFVVGAL AGMRLVGLLRQDWEYLGYARRARGVADHARLRRFIGQAFALLVFALRRGSKLATAMEARGFGAYPTRTWA RPSTFGVPEMVLIAGAVVIAAAAVAVSTATGHWDFIVGR
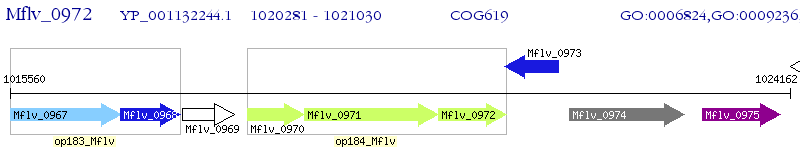
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0972 | - | - | 100% (249) | cobalt transport protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_6724 | - | 2e-98 | 73.88% (245) | ABC-type cobalt transport system |
| M. thermoresistible (build 8) | TH_3324 | - | 3e-05 | 31.44% (229) | PUTATIVE - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0972|M.gilvum_PYR-GCK ------------MIRVNPVAKLLAA-LVIAGVLVLSVDWVSALTALVLEI
MSMEG_6724|M.smegmatis_MC2_155 -MSITVDGAPIRRVGINPVAKLAAA-LVIALGLVLSVDWVSAATALVLEI
TH_3324|M.thermoresistible__bu VNPLDLSAAHNRWSGVPAAEKLCFYGGLLLCAMLLPPRTGAPLVLLVVAV
: .. ** :: ::*. :. . **: :
Mflv_0972|M.gilvum_PYR-GCK PLFLAMRIPLRAFLIRGAFITLAAALTALTNLLYGEPGGAVYWHFLLVTV
MSMEG_6724|M.smegmatis_MC2_155 VLMLVLKVPVRTLLTRGALVLLAAALTGVTILLYGEQSGTVHWHFLLITV
TH_3324|M.thermoresistible__bu STLVFARVQVRLFLLAMLGPLVFIMLGALPIAVS--LRGGPHWE------
:: :: :* :* : * .:. : * :*.
Mflv_0972|M.gilvum_PYR-GCK SDGSVELAAAMFLRVLAIALPSVCLFIDVSPTELADGLGQIMRLPARFVV
MSMEG_6724|M.smegmatis_MC2_155 SDGSISLALATFLRVLAIALPSVILFIDTDPTELADGLGQVLRLPARFVL
TH_3324|M.thermoresistible__bu -PGGVDLALDTALRALAASAATISLAMTTLMADLLDLARRAGTPPALCHV
*.:.** **.** : .:: * : . ::* * : ** :
Mflv_0972|M.gilvum_PYR-GCK GALAGMRLVG-LLRQDWEYLGYARRARGVADHARLRRFIGQAFALLVF-A
MSMEG_6724|M.smegmatis_MC2_155 GALAGLRMVG-LLGQDWRYLGYARRARGVADQDRLRRFAAQGFALLVS-A
TH_3324|M.thermoresistible__bu ADLT-YRLVGILLRSALRAREAVGLRLGLRRPRDAITVIGTQFALIFIRG
. *: *:** ** . . . *: . . ***:. .
Mflv_0972|M.gilvum_PYR-GCK LRRGSKLATAMEARGFGAYPTRTWARPSTFGVPEMVLIAGAVVIAAAAVA
MSMEG_6724|M.smegmatis_MC2_155 VRRGSMLATAMEARGFGAHPRRTWARPSRFGMRESALILAAFVIAAAAIS
TH_3324|M.thermoresistible__bu TDRARAMSEAMSLR---ADPGATAVLTSARPVRASRLVGTAALLAAIAAV
*. :: **. * * * * . .* : *: * ::** *
Mflv_0972|M.gilvum_PYR-GCK VSTATGHWDFIVGR
MSMEG_6724|M.smegmatis_MC2_155 VSVATGHWNFIGHR
TH_3324|M.thermoresistible__bu ATLTWLGWKGWHGV
.: : *.