For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSDVATQIAFVQATWHRNIVDQARAGFTDRINGLGFSSDTLEFFEVPGAFEIPLTAKRLAKTGRYRAIVA AGLVVDGGIYRHEFVATAVIDGLMRVQLDTDVPVFSVVLTPHHFHEHDEHVRYFAEHFVKKGAEAANAVA ATLELHASLG
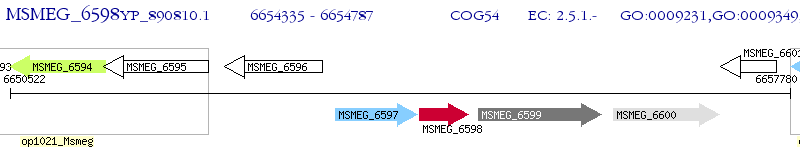
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6598 | - | - | 100% (150) | riboflavin synthase subunit beta |
| M. smegmatis MC2 155 | MSMEG_3073 | ribH | 1e-11 | 36.23% (138) | riboflavin synthase subunit beta |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1451 | ribH | 2e-11 | 33.09% (136) | 6,7-dimethyl-8-ribityllumazine synthase |
| M. gilvum PYR-GCK | Mflv_3718 | ribH | 6e-12 | 34.07% (135) | riboflavin synthase subunit beta |
| M. tuberculosis H37Rv | Rv1416 | ribH | 2e-11 | 33.09% (136) | 6,7-dimethyl-8-ribityllumazine synthase |
| M. leprae Br4923 | MLBr_00560 | ribH | 2e-11 | 32.35% (136) | 6,7-dimethyl-8-ribityllumazine synthase |
| M. abscessus ATCC 19977 | MAB_2795c | - | 5e-11 | 34.81% (135) | 6,7-dimethyl-8-ribityllumazine synthase |
| M. marinum M | MMAR_2223 | ribH | 5e-12 | 39.45% (109) | riboflavin synthase beta chain RibH |
| M. avium 104 | MAV_3364 | ribH | 7e-12 | 34.29% (140) | riboflavin synthase subunit beta |
| M. thermoresistible (build 8) | TH_1012 | ribH | 5e-11 | 36.54% (104) | PROBABLE RIBOFLAVIN SYNTHASE BETA CHAIN RIBH |
| M. ulcerans Agy99 | MUL_1810 | ribH | 1e-11 | 37.50% (104) | 6,7-dimethyl-8-ribityllumazine synthase |
| M. vanbaalenii PYR-1 | Mvan_2695 | ribH | 7e-14 | 36.03% (136) | riboflavin synthase subunit beta |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00560|M.leprae_Br4923 MSGGAGIPEVPGIDASGLRLGIVASTWHSRICDALLAGARKVAADSGIDG
MAV_3364|M.avium_104 MSPAAGVPEMPALDASGVRLGIVASTWHSRICDALLAGARKVAADSGVEN
Mb1451|M.bovis_AF2122/97 ------MPDLPSLDASGVRLAIVASSWHGKICDALLDGARKVAAGCGLDD
Rv1416|M.tuberculosis_H37Rv ------MPDLPSLDASGVRLAIVASSWHGKICDALLDGARKVAAGCGLDD
MMAR_2223|M.marinum_M MSGGAGIPDVPAFDASDVRLAIVASTWHTKICDALLAGARNTAADSGIDN
MUL_1810|M.ulcerans_Agy99 MSGGAGIPDVPAFDASGVRLAIVASTWHTKICDALLAGARNTAADSGIDN
Mflv_3718|M.gilvum_PYR-GCK MS-GAGVPDMPALHAEDVTLAIVASTWHETICDALLDGARKVAEQAGVTD
Mvan_2695|M.vanbaalenii_PYR-1 MSPAAGVPELPALDGKGLRLAIVASTWHETICDALLDGARKVAVGAGIDD
TH_1012|M.thermoresistible__bu VS-KAGVPDLPQLDASGLKLAIVASTWHQKICDALLDGARKVAADAGITD
MAB_2795c|M.abscessus_ATCC_199 MS-GDGIPSLSVGDASGISLAIVASTWHDQICTALLEGAQRVATEAGIDR
MSMEG_6598|M.smegmatis_MC2_155 ------------MSDVATQIAFVQATWHRNIVDQARAGFTDRINGLGFSS
:.:* ::** * * *.
MLBr_00560|M.leprae_Br4923 P--TVVRVLGAIEIPVVVQELARH--HDAVVALGVVIRGDTPHFDYVCNS
MAV_3364|M.avium_104 P--TVVRVLGAIEIPVVAQELARN--HDAVVALGVVIRGQTPHFEYVCDA
Mb1451|M.bovis_AF2122/97 P--TVVRVLGAIEIPVVAQELARN--HDAVVALGVVIRGQTPHFDYVCDA
Rv1416|M.tuberculosis_H37Rv P--TVVRVLGAIEIPVVAQELARN--HDAVVALGVVIRGQTPHFDYVCDA
MMAR_2223|M.marinum_M P--TVVRVLGAIEIPVVAQELTRN--HDAVVALGVVIRGETPHFDYVCDA
MUL_1810|M.ulcerans_Agy99 P--TVVRVLGAIEIPVVAQELTRN--HDAVVALGVVIRGETPHFDYVCDV
Mflv_3718|M.gilvum_PYR-GCK P--TVVRVLGAIEIPVVAQALART--HDAVIALGVVIRGETPHFDYVCDA
Mvan_2695|M.vanbaalenii_PYR-1 P--TVVRVLGAIEIPVVAQALART--HDAVVALGVVIRGETPHFDYVCDA
TH_1012|M.thermoresistible__bu P--TVVRVLGAIEIPVVAQALAAN--HDAVIALGVVIRGETPHFDYVCDA
MAB_2795c|M.abscessus_ATCC_199 P--TVVRVLGAIEIPVVAQALART--HDAVVALGVVIQGETPHFGYVCDA
MSMEG_6598|M.smegmatis_MC2_155 DTLEFFEVPGAFEIPLTAKRLAKTGRYRAIVAAGLVVDGGIYRHEFVATA
...* **:***:..: *: : *::* *:*: * :. :*.
MLBr_00560|M.leprae_Br4923 VTQGLTRIALDTSTPVGNGVLTTNTEKQALDRAGLPTSAEDKGAQAAAAA
MAV_3364|M.avium_104 VTQGITRVSLDASTPVANGVLTTDNEQQALDRAGLPDSAEDKGAQAAGAA
Mb1451|M.bovis_AF2122/97 VTQGLTRVSLDSSTPIANGVLTTNTEEQALDRAGLPTSAEDKGAQATVAA
Rv1416|M.tuberculosis_H37Rv VTQGLTRVSLDSSTPIANGVLTTNTEEQALDRAGLPTSAEDKGAQATVAA
MMAR_2223|M.marinum_M VTQGLTRVSLDSSTPVANGVLTTNSEEQALNRAGLPTSDEDKGAQATAAA
MUL_1810|M.ulcerans_Agy99 VTQGLTRVSLDSSTPVANGVLTTNSEEQALNRAGLPTSDEDKGAQATAAA
Mflv_3718|M.gilvum_PYR-GCK VTQGLTRVSLDASTPVANGVLTVNTENQALDRAGLPNSAEDKGAQAAAAA
Mvan_2695|M.vanbaalenii_PYR-1 VTQGLTRVSLDASTPVANGVLTTNTEHQALDRAGLPDSAEDKGAQAAAAA
TH_1012|M.thermoresistible__bu VTQGLTRVSLDASTPVANGVLTTDTEAQALDRAGLPGSAEDKGGQAAAAA
MAB_2795c|M.abscessus_ATCC_199 VTTGLTRVSLDTSTPVANGVLTVNNEQQAIDRAGLPGSAEDKGAQAAAAA
MSMEG_6598|M.smegmatis_MC2_155 VIDGLMRVQLDTDVPVFSVVLTPHHFHEHDEHVRYFAEHFVKKGAEAANA
* *: *: **:..*: . *** . : ::. . * . : *
MLBr_00560|M.leprae_Br4923 LTTALTLLNLRSRI-
MAV_3364|M.avium_104 LSAALTLRELRARS-
Mb1451|M.bovis_AF2122/97 LATALTLRELRAHS-
Rv1416|M.tuberculosis_H37Rv LATALTLRELRAHS-
MMAR_2223|M.marinum_M LTTALTLRELRAEA-
MUL_1810|M.ulcerans_Agy99 LTTALTLRELRAES-
Mflv_3718|M.gilvum_PYR-GCK LSTALTLRQLASPS-
Mvan_2695|M.vanbaalenii_PYR-1 LSTALTLRQLS----
TH_1012|M.thermoresistible__bu LSTALTLRELRSQI-
MAB_2795c|M.abscessus_ATCC_199 LDTALTLRRLRQPWA
MSMEG_6598|M.smegmatis_MC2_155 VAATLELHASLG---
: ::* *