For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VPDLPSLDASGVRLAIVASSWHGKICDALLDGARKVAAGCGLDDPTVVRVLGAIEIPVVAQELARNHDAV VALGVVIRGQTPHFDYVCDAVTQGLTRVSLDSSTPIANGVLTTNTEEQALDRAGLPTSAEDKGAQATVAA LATALTLRELRAHS
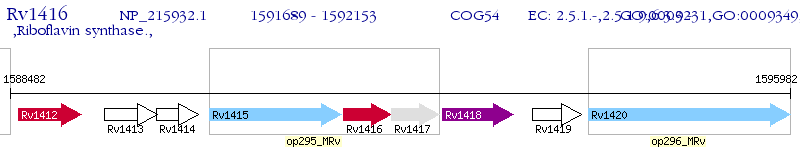
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1416 | ribH | - | 100% (154) | riboflavin synthase subunit beta |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1451 | ribH | 2e-83 | 100.00% (154) | 6,7-dimethyl-8-ribityllumazine synthase |
| M. gilvum PYR-GCK | Mflv_3718 | ribH | 3e-66 | 80.52% (154) | riboflavin synthase subunit beta |
| M. leprae Br4923 | MLBr_00560 | ribH | 6e-67 | 78.95% (152) | 6,7-dimethyl-8-ribityllumazine synthase |
| M. abscessus ATCC 19977 | MAB_2795c | - | 6e-61 | 76.16% (151) | 6,7-dimethyl-8-ribityllumazine synthase |
| M. marinum M | MMAR_2223 | ribH | 3e-70 | 84.42% (154) | riboflavin synthase beta chain RibH |
| M. avium 104 | MAV_3364 | ribH | 2e-69 | 83.12% (154) | riboflavin synthase subunit beta |
| M. smegmatis MC2 155 | MSMEG_3073 | ribH | 5e-66 | 81.46% (151) | riboflavin synthase subunit beta |
| M. thermoresistible (build 8) | TH_1012 | ribH | 5e-70 | 84.87% (152) | PROBABLE RIBOFLAVIN SYNTHASE BETA CHAIN RIBH |
| M. ulcerans Agy99 | MUL_1810 | ribH | 4e-71 | 85.06% (154) | 6,7-dimethyl-8-ribityllumazine synthase |
| M. vanbaalenii PYR-1 | Mvan_2695 | ribH | 9e-70 | 85.33% (150) | riboflavin synthase subunit beta |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3718|M.gilvum_PYR-GCK MS-GAGVPDMPALHAEDVTLAIVASTWHETICDALLDGARKVAEQAGVTD
Mvan_2695|M.vanbaalenii_PYR-1 MSPAAGVPELPALDGKGLRLAIVASTWHETICDALLDGARKVAVGAGIDD
Rv1416|M.tuberculosis_H37Rv ------MPDLPSLDASGVRLAIVASSWHGKICDALLDGARKVAAGCGLDD
Mb1451|M.bovis_AF2122/97 ------MPDLPSLDASGVRLAIVASSWHGKICDALLDGARKVAAGCGLDD
MMAR_2223|M.marinum_M MSGGAGIPDVPAFDASDVRLAIVASTWHTKICDALLAGARNTAADSGIDN
MUL_1810|M.ulcerans_Agy99 MSGGAGIPDVPAFDASGVRLAIVASTWHTKICDALLAGARNTAADSGIDN
MLBr_00560|M.leprae_Br4923 MSGGAGIPEVPGIDASGLRLGIVASTWHSRICDALLAGARKVAADSGIDG
MAV_3364|M.avium_104 MSPAAGVPEMPALDASGVRLGIVASTWHSRICDALLAGARKVAADSGVEN
TH_1012|M.thermoresistible__bu -VSKAGVPDLPQLDASGLKLAIVASTWHQKICDALLDGARKVAADAGITD
MAB_2795c|M.abscessus_ATCC_199 -MSGDGIPSLSVGDASGISLAIVASTWHDQICTALLEGAQRVATEAGIDR
MSMEG_3073|M.smegmatis_MC2_155 -MSGVGVPDLPEIDASSLTLAIVASTWHTEICDALLEGARKVAADAGIPD
:*.:. ....: *.****:** ** *** **:..* .*:
Mflv_3718|M.gilvum_PYR-GCK PTVVRVLGAIEIPVVAQALARTHDAVIALGVVIRGETPHFDYVCDAVTQG
Mvan_2695|M.vanbaalenii_PYR-1 PTVVRVLGAIEIPVVAQALARTHDAVVALGVVIRGETPHFDYVCDAVTQG
Rv1416|M.tuberculosis_H37Rv PTVVRVLGAIEIPVVAQELARNHDAVVALGVVIRGQTPHFDYVCDAVTQG
Mb1451|M.bovis_AF2122/97 PTVVRVLGAIEIPVVAQELARNHDAVVALGVVIRGQTPHFDYVCDAVTQG
MMAR_2223|M.marinum_M PTVVRVLGAIEIPVVAQELTRNHDAVVALGVVIRGETPHFDYVCDAVTQG
MUL_1810|M.ulcerans_Agy99 PTVVRVLGAIEIPVVAQELTRNHDAVVALGVVIRGETPHFDYVCDVVTQG
MLBr_00560|M.leprae_Br4923 PTVVRVLGAIEIPVVVQELARHHDAVVALGVVIRGDTPHFDYVCNSVTQG
MAV_3364|M.avium_104 PTVVRVLGAIEIPVVAQELARNHDAVVALGVVIRGQTPHFEYVCDAVTQG
TH_1012|M.thermoresistible__bu PTVVRVLGAIEIPVVAQALAANHDAVIALGVVIRGETPHFDYVCDAVTQG
MAB_2795c|M.abscessus_ATCC_199 PTVVRVLGAIEIPVVAQALARTHDAVVALGVVIQGETPHFGYVCDAVTTG
MSMEG_3073|M.smegmatis_MC2_155 PTVVRVLGAIEIPVVAQALARTHDAVVALGVVIQGETPHFDYVCDAVTQG
***************.* *: ****:******:*:**** ***: ** *
Mflv_3718|M.gilvum_PYR-GCK LTRVSLDASTPVANGVLTVNTENQALDRAGLPNSAEDKGAQAAAAALSTA
Mvan_2695|M.vanbaalenii_PYR-1 LTRVSLDASTPVANGVLTTNTEHQALDRAGLPDSAEDKGAQAAAAALSTA
Rv1416|M.tuberculosis_H37Rv LTRVSLDSSTPIANGVLTTNTEEQALDRAGLPTSAEDKGAQATVAALATA
Mb1451|M.bovis_AF2122/97 LTRVSLDSSTPIANGVLTTNTEEQALDRAGLPTSAEDKGAQATVAALATA
MMAR_2223|M.marinum_M LTRVSLDSSTPVANGVLTTNSEEQALNRAGLPTSDEDKGAQATAAALTTA
MUL_1810|M.ulcerans_Agy99 LTRVSLDSSTPVANGVLTTNSEEQALNRAGLPTSDEDKGAQATAAALTTA
MLBr_00560|M.leprae_Br4923 LTRIALDTSTPVGNGVLTTNTEKQALDRAGLPTSAEDKGAQAAAAALTTA
MAV_3364|M.avium_104 ITRVSLDASTPVANGVLTTDNEQQALDRAGLPDSAEDKGAQAAGAALSAA
TH_1012|M.thermoresistible__bu LTRVSLDASTPVANGVLTTDTEAQALDRAGLPGSAEDKGGQAAAAALSTA
MAB_2795c|M.abscessus_ATCC_199 LTRVSLDTSTPVANGVLTVNNEQQAIDRAGLPGSAEDKGAQAAAAALDTA
MSMEG_3073|M.smegmatis_MC2_155 LTRVSLDEATPVANGVLTTNTEEQARARAGLPGSTEDKGAQAAAAALSTA
:**::** :**:.*****.:.* ** ***** * ****.**: *** :*
Mflv_3718|M.gilvum_PYR-GCK LTLRQLASPS-
Mvan_2695|M.vanbaalenii_PYR-1 LTLRQLS----
Rv1416|M.tuberculosis_H37Rv LTLRELRAHS-
Mb1451|M.bovis_AF2122/97 LTLRELRAHS-
MMAR_2223|M.marinum_M LTLRELRAEA-
MUL_1810|M.ulcerans_Agy99 LTLRELRAES-
MLBr_00560|M.leprae_Br4923 LTLLNLRSRI-
MAV_3364|M.avium_104 LTLRELRARS-
TH_1012|M.thermoresistible__bu LTLRELRSQI-
MAB_2795c|M.abscessus_ATCC_199 LTLRRLRQPWA
MSMEG_3073|M.smegmatis_MC2_155 VTLRELRRA--
:** .*