For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SERQELRELRARLDGLTIRDAARLGRRLKNLRGDDLTKNFAKIEQQIAQAEALVLTRQAAVPQITYPDLP VTQRRDDLAKAIAENQVVVVAGATGSGKTTQLPKICLELGRGIRGTIGHTQPRRLAARTVAQRIADELGT PLGEAVGYTVRFTDQASDSTLVKLMTDGILLAEIQRDRRLLRYDTLILDEAHERSLNIDFLLGYLRELLP RRPDLKVIVTSATIEPERFSRHFGDAPIVEVSGRTYPVEIRYRPLEVPAPTSDDEDPDDPDHEIVRTEVR DPTEAIVDAVRELEAEPPGDVLVFLSGEREIRDTAEALKDLRNTEVLPLYARLPTAEQQKVFQPSHAGRR VVLATNVAETSLTVPGIRYVVDPGTARISRYSRRTKVQRLPIEPISQASAAQRAGRSGRTAPGVCIRLYS EEDFESRPRYTDPEILRTNLAAVILQMSALGLGDIENFPFLDPPDARSIRDGIQLLQELGAFDTSSAAGG GNSASAAGGGNSVPALTETGRRLARLPLDPRIGRMILQADAEGCVREALVLAAALSIPDPRERPSDREEA ARQKHARFADEHSDFISYLNLWNYLREQRNELSGNAFRRMCRDEFLHYLRIREWQDLVGQLRSIARDIGI RESDEPADPARIHAALTAGLLSHVGLREGDTRDYTGARNTKFVLAPGSVLTKKPPRWIVVADLVETSRLF GRIAARIEPEAVERVAGHLVQRTYSEPHWDARRGAVMASERVTLYGLPLVPRRRVNYAQIDPVVSRELFI RHALVEGEWQTRHHFFRDNARLREEVEEMEERARRRDLLVGDDEIFALYDARIPADVVSARHFDAWWKKQ RHKTPDLLTFTREDLLRTDDAADHPDTWRAGDLELPLSYRYQPGAADDGVTVHVPVGVLARLGGENFAWQ VPALREELVTALIKSLPKDLRRNFVPAPDTARAILPHLDPGTGSLLDAVQRELRRRTGILVPVDAFDLDK VPPHLRVTFAVENSDGAEVARGKDIETLREQLAVPVAQAVAEAVDDGLQRTGLRAWPDDLDELPRTVENV SGGHTVRGYPGFVDTGKAVDIRVFANPIERDAAMGPGLRRLLRLSVPSPVKAIERGLGTRTRLALNANPD GNLAALLEDCADAAADTLVRQIVWTRADFTVLVQRAAKELGATTQAAVTRVEKVLAAAHEVGVALPDKPP AAQVDAVEDIRLQLERLLPKGFVTATGVARLTDLTRYVTAIARRLERLPHAIGADRERMARVHAVEDAYD DLVRALSPARAAAHDVSAIAWQIEELRVSLWAQQLGTPRPVSEQRIYKAIDAVAR*
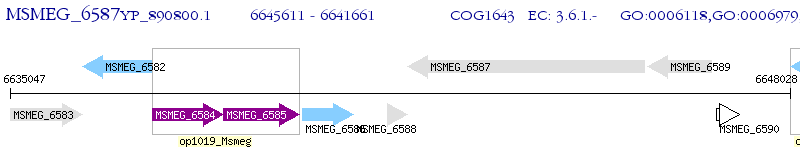
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6587 | hrpA | - | 100% (1316) | ATP-dependent helicase HrpA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1063 | - | 0.0 | 75.91% (1324) | ATP-dependent helicase HrpA |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0056c | - | 0.0 | 77.52% (1312) | ATP-dependent helicase HrpA |
| M. marinum M | MMAR_2947 | - | 0.0 | 73.90% (1318) | HrpA-like helicase |
| M. avium 104 | MAV_2737 | hrpA | 0.0 | 76.38% (1308) | ATP-dependent helicase HrpA |
| M. thermoresistible (build 8) | TH_0802 | hrpA | 0.0 | 75.13% (1327) | ATP-dependent helicase HrpA |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5766 | - | 0.0 | 77.64% (1315) | ATP-dependent helicase HrpA |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6587|M.smegmatis_MC2_155 ------MSERQELRELRARLDGLTIRDAARLGRRLKNLRGDDLTKNFAKI
MAB_0056c|M.abscessus_ATCC_199 ------MSELS-RTELRARLDGVTFRDASRLGRRLKNQR-DTNPDALTKL
Mflv_1063|M.gilvum_PYR-GCK ----MPEQPSTDVRALRARLDGLTISDAARLGRRLRQQR-TPDPAALARL
Mvan_5766|M.vanbaalenii_PYR-1 ----MPEPSRTDVRALRARLDDLTISDAARLGRRLRQLR-DPSEEQLGKL
TH_0802|M.thermoresistible__bu -----VSDPSHDVRALRARLDGLTHRDAARLGRRLRQVR-GAKPEQLQRI
MMAR_2947|M.marinum_M -------MADLSVAQLRSRLDGLSTRDAVRLGRRLKGLR-GAKPERLQQL
MAV_2737|M.avium_104 MAAVAEPSFPVSGAQLRGRLDGLTIRDAARLGRRLKKLR-GAAPDKLRQL
**.***.:: ** ******: * : ::
MSMEG_6587|M.smegmatis_MC2_155 EQ----QIAQAEALVLTRQAAVPQITYPDLPVTQRRDDLAKAIAENQVVV
MAB_0056c|M.abscessus_ATCC_199 AE----QITAAEALIATRAAAVPVITYPDLPVSAHREELAHAISAHQVVV
Mflv_1063|M.gilvum_PYR-GCK AE----RFTAAEALVATRLAAVPEITYPDLPVSERRDDIAAAIAANQVVI
Mvan_5766|M.vanbaalenii_PYR-1 AK----QFDTAEALVASRLAAVPQISYPDLPVTERRDEIAAAIAANQVVI
TH_0802|M.thermoresistible__bu AE----QIAAGEALVAARAAAVPEISYPDLPVSERRHEIAEAIANHQVVV
MMAR_2947|M.marinum_M TEQIAVQISAAEALIATRQAAVPTITYPDLPVSERRQEIAEAIRANQVVV
MAV_2737|M.avium_104 AD----QIAAAEAVVAARHAAVPAVSYPDLPVSERRREIADAIRAHQVVV
. :: .**:: :* **** ::******: :* ::* ** :***:
MSMEG_6587|M.smegmatis_MC2_155 VAGATGSGKTTQLPKICLELGRGIRGTIGHTQPRRLAARTVAQRIADELG
MAB_0056c|M.abscessus_ATCC_199 VAGATGSGKTTQLPKICLELGRGIRGTIGHTQPRRLAARTVAERIAEELG
Mflv_1063|M.gilvum_PYR-GCK VAGETGSGKTTQLPKICLEMGRGIRGTIGHTQPRRLAARTVATRIAEELG
Mvan_5766|M.vanbaalenii_PYR-1 VAGETGSGKTTQLPKICLELGRGIRGTIGHTQPRRLAARTVAARIAEELG
TH_0802|M.thermoresistible__bu VAGETGSGKTTQLPKICLELGRGIRGTIGHTQPRRLAARTVAQRIADELG
MMAR_2947|M.marinum_M VAGETGSGKTTQLPKICLDIGRGVRGTIGHTQPRRLAARTVAQRIADELQ
MAV_2737|M.avium_104 IAGETGSGKTTQLPKVCLEAGRGIRGTIGHTQPRRLAARTVAQRIADELG
:** ***********:**: ***:****************** ***:**
MSMEG_6587|M.smegmatis_MC2_155 TPLGEAVGYTVRFTDQASDSTLVKLMTDGILLAEIQRDRRLLRYDTLILD
MAB_0056c|M.abscessus_ATCC_199 TQLGETVGYTVRFTDQASDRTLVKLMTDGILLAEIQRDRRLLRYDTLILD
Mflv_1063|M.gilvum_PYR-GCK TPLGEAVGYTVRFTDQASDRTLVKLMTDGILLAEVQRDRRLLRYDTLIID
Mvan_5766|M.vanbaalenii_PYR-1 TPLGEAVGYTVRFTDQASDRTLVKLMTDGILLAEVQRDRRLLRYDTLIID
TH_0802|M.thermoresistible__bu TPLGEAVGYTVRFTDQAGDRTLVKLMTDGILLAEIQRDRRLLRYDTIIID
MMAR_2947|M.marinum_M TPLGDAVGYAVRFTDQVSERTLVKLMTDGILLAEIQRDRRLLRYDTLILD
MAV_2737|M.avium_104 SPLGETVGYTVRFTDQVSDRTLIKLMTDGILLAEIQRDRRLLRYDTLILD
: **::***:******..: **:***********:***********:*:*
MSMEG_6587|M.smegmatis_MC2_155 EAHERSLNIDFLLGYLRELLPRRPDLKVIVTSATIEPERFSRHFGD----
MAB_0056c|M.abscessus_ATCC_199 EAHERSLNIDFLLGYLRELLPRRPDLKIIVTSATIEPQRFSAHFGN----
Mflv_1063|M.gilvum_PYR-GCK EAHERSLNIDFLLGYLRRLLPRRPDLKVIVTSATIEPERFARFFAGGEHG
Mvan_5766|M.vanbaalenii_PYR-1 EAHERSLNIDFLLGYLRQLLPRRPDLKVIVTSATIEPERFAAHFAG----
TH_0802|M.thermoresistible__bu EAHERSLNIDFLLGYLKGLLPRRPDLKVIITSATIEPRRFAKHFDA----
MMAR_2947|M.marinum_M EAHERSLNIDFLLGYLRELLPRRPDLKLIVTSATIEPQRFAAHFGG----
MAV_2737|M.avium_104 EAHERSLNIDFLLGYLRALLPRRPDLKLIITSATIEPRRFSEHFAN----
****************: *********:*:*******.**: .*
MSMEG_6587|M.smegmatis_MC2_155 ----APIVEVSGRTYPVEIRYRPLEVPAPTSD----DEDPDDPDHEIVRT
MAB_0056c|M.abscessus_ATCC_199 ----APIVEVSGRTYPVEIRYRPLEVPVHGTSGAKAEDEPDDPDHEIVRT
Mflv_1063|M.gilvum_PYR-GCK SGAGAPIVEVSGRTYPVEIRHRPLEVAVSAAD----DDDPDDPDHEVVRT
Mvan_5766|M.vanbaalenii_PYR-1 ----APIVEVSGRTYPVEIRYRPLEVTVPGQD----GEDPDDPDHEVVRT
TH_0802|M.thermoresistible__bu -----PVVEVSGRTYPVEIRYRPLEVPAGTDT----ADDPDDPDHEIVRT
MMAR_2947|M.marinum_M ----APIIEVSGRTYPVAIRYRPLEVPVSTGSPED-SSDPDDPDHEIVRT
MAV_2737|M.avium_104 ----APVIEVSGRTYPVEIRYRPLEIALPSATAD----DPDDPDHEIVRT
*::********* **:****:. :*******:***
MSMEG_6587|M.smegmatis_MC2_155 EVRDPTEAIVDAVRELEAEPPGDVLVFLSGEREIRDTAEALKDLR---NT
MAB_0056c|M.abscessus_ATCC_199 ELRDQTEAIADAVRELQNEPPGDVLVFLSGEREIRDTSEALRDIVRA-GT
Mflv_1063|M.gilvum_PYR-GCK ELRDPTEAIIDAVAELESEPRGDVLVFLSGEREIRDTAEALRAVTDPGHT
Mvan_5766|M.vanbaalenii_PYR-1 ELRDPTEAIIDAVAELEAEPPGDVLVFLSGEREIRDTAEALRAVVDPGHT
TH_0802|M.thermoresistible__bu EIRDQTEAIIDAVRELDAEPPGDVLVFLSGEREIRDTAEALRGTLDP-DV
MMAR_2947|M.marinum_M ETRDEVEAIVDAVHELQNEAPGDVLVFLSGEREIRDTAEALSGLE---HT
MAV_2737|M.avium_104 ETRDEVEAIVDAIAELEAEPPGDVLVFLSGEREIRDTAEALTGLK---HT
* ** .*** **: **: *. ****************:*** .
MSMEG_6587|M.smegmatis_MC2_155 EVLPLYARLPTAEQQKVFQPSHAGRRVVLATNVAETSLTVPGIRYVVDPG
MAB_0056c|M.abscessus_ATCC_199 EVLPLYARLPTAEQHKVFA-SHTGRRVVLATNVAETSLTVPGIRYVIDPG
Mflv_1063|M.gilvum_PYR-GCK EVLPLYARLPTAEQQKVFSPSRAARRIVLATNVAETSLTVPGIRYVVDPG
Mvan_5766|M.vanbaalenii_PYR-1 EVLPLYARLPTAEQQKVFHPGRTARRIVLATNVAETSLTVPGIRYVVDPG
TH_0802|M.thermoresistible__bu EVLPLYARLPTAEQQKVFTPHRLRRRIVLATNVAETSLTVPGIRYVVDPG
MMAR_2947|M.marinum_M EVLPLYARLPTAEQQKVFA-PHTGRRVVLATNVAETSLTVPGIRYVVDPG
MAV_2737|M.avium_104 EVLPLYARLPTAEQQKVFA-PHTGRRVVLATNVAETSLTVPGIRYVVDPG
**************:*** : **:*******************:***
MSMEG_6587|M.smegmatis_MC2_155 TARISRYSRRTKVQRLPIEPISQASAAQRAGRSGRTAPGVCIRLYSEEDF
MAB_0056c|M.abscessus_ATCC_199 TARISRYSRRTKVQRLPIEPISQASAAQRSGRSGRTAPGICIRLYSEQDF
Mflv_1063|M.gilvum_PYR-GCK TARISRYSRRTKVQRLPIEPISQASAAQRAGRSGRTAPGVCIRLYSEQDF
Mvan_5766|M.vanbaalenii_PYR-1 TARISRYSRRTKVQRLPIEPISQASAAQRAGRSGRTAPGVCIRLYSEQDF
TH_0802|M.thermoresistible__bu TARISRYSRRTKVQRLPIEPISQASAAQRAGRSGRVAPGVCIRLYSEEDF
MMAR_2947|M.marinum_M NARISRYSRRLKVQRLPIEPISQASAAQRAGRCGRVAPGICIRLYSEDDF
MAV_2737|M.avium_104 NARISRYSRRLKVQRLPIEPISQASAAQRAGRCGRVAPGVCIRLYSEADF
.********* ******************:**.**.***:******* **
MSMEG_6587|M.smegmatis_MC2_155 ESRPRYTDPEILRTNLAAVILQMSALGLGDIENFPFLDPPDARSIRDGIQ
MAB_0056c|M.abscessus_ATCC_199 EARPRYTDPEILRTNLAAVILQMAALQLGDIENFPFLDPPDKRSIRDGVQ
Mflv_1063|M.gilvum_PYR-GCK ESRPRYTDPEILRTNLAAVILQMAALGFGDIEAFGFLDPPDARSIRDGVA
Mvan_5766|M.vanbaalenii_PYR-1 EARPRYTDPEILRTNLAAVILQMAALGFGDIEGFGFLDPPDARSIRDGVA
TH_0802|M.thermoresistible__bu ESRPEFTEPEILRTNLAAVILQMAALGFGAVEDFPFLDPPDQRSIRDGVA
MMAR_2947|M.marinum_M AARPQYTDPEILRTNLAAVILQMAALQLGEIENFGFLDPPDRRSVRDGVQ
MAV_2737|M.avium_104 AARPRYTEPEILRTNLAAVLLQMAALQLGDIENFPFLDPPDRRSVRDGVQ
:**.:*:***********:***:** :* :* * ****** **:***:
MSMEG_6587|M.smegmatis_MC2_155 LLQELGAFDTSSAAGGGNSASAAGGGNSVPALTETGRRLARLPLDPRIGR
MAB_0056c|M.abscessus_ATCC_199 LLQELGAFSADG------------------TITELGRRLARLPLDPRIGR
Mflv_1063|M.gilvum_PYR-GCK LLQELGAFDRQG------------------ELTDIGRRLAQIPVDPRLGR
Mvan_5766|M.vanbaalenii_PYR-1 LLQELGAFDQQA------------------DLTDIGRRLAQIPVDPRLGR
TH_0802|M.thermoresistible__bu LLRELGAFDERG------------------EITDVGRRLAQLPVDPRLGR
MMAR_2947|M.marinum_M LLQELGAFDPTG------------------AITDLGRRLARLPVDPRLGR
MAV_2737|M.avium_104 LLTELGAFDRQG------------------AITERGRRLARLPVDPRLGR
** *****. . :*: *****::*:***:**
MSMEG_6587|M.smegmatis_MC2_155 MILQADAEGCVREALVLAAALSIPDPRERPSDREEAARQKHARFADEHSD
MAB_0056c|M.abscessus_ATCC_199 MILQADAEGCVEEILVLAAALSIPDPRERPADREEAARQKHARFADDHSD
Mflv_1063|M.gilvum_PYR-GCK MILQADHEGCVREMLVLAAALSIPDPRERPADREDAARQKHARFADEHSD
Mvan_5766|M.vanbaalenii_PYR-1 MILQADAEGCVREMLVLAAALSIPDPRERPADKEEAARQKHARFADQHSD
TH_0802|M.thermoresistible__bu MVLQAETEGCVREVLVLAAALSIPDPRERPLDKEDAARAKHARFADEHSD
MMAR_2947|M.marinum_M MILQAGVEGCLREVLVLAAALTIPDPRERPTDREEAARQKHARFADEDSD
MAV_2737|M.avium_104 MILAAQTEGCVREMLVLAAALSIPDPRERPSDREEAARQKHARFADEHSD
*:* * ***:.* *******:******** *:*:*** *******:.**
MSMEG_6587|M.smegmatis_MC2_155 FISYLNLWNYLREQRNELSGNAFRRMCRDEFLHYLRIREWQDLVGQLRSI
MAB_0056c|M.abscessus_ATCC_199 FIAYLNLWRYLQDQRSQLSGSAFRRMCRNEFLHYLRIREWQDLVGQLRSI
Mflv_1063|M.gilvum_PYR-GCK FTSFLNLWWYLTEQRKERSGSSFRRMCRDEFLHYLRIREWQDLVGQLRSI
Mvan_5766|M.vanbaalenii_PYR-1 FTSYLNLWHYLTEQRKERSGSSFRRMCRDEFLHYLRIREWQDLVGQLRGI
TH_0802|M.thermoresistible__bu FMSYLNLWRYLTEQRKERSGNSFRRMCREEFLHYLRIREWQDLVGQLRSI
MMAR_2947|M.marinum_M FMSYLNLWRYLGEQRRALSRSAFRRMCRNDFLHYLRIREWQDLVGQLRSI
MAV_2737|M.avium_104 FMSYLNLWRYLREQRKELSGNQFRRLCRAEFLHYLRIREWQDLVGQLRGI
* ::**** ** :** * . ***:** :******************.*
MSMEG_6587|M.smegmatis_MC2_155 ARDIGIRE----SDEPADPARIHAALTAGLLSHVGLREGDT---------
MAB_0056c|M.abscessus_ATCC_199 ARDLGIRE----SGERADPGLVHAALIAGLLSHIGLREGDT---------
Mflv_1063|M.gilvum_PYR-GCK CRDLGIRE----QDEPADPRSVHAALTAGLLSHVGMRDTDG---------
Mvan_5766|M.vanbaalenii_PYR-1 CRDIGIRE----QDEPADPAAVHAALAAGLLSHVGMRDTDG---------
TH_0802|M.thermoresistible__bu ARDIGIRES--PDPEPAAPDRIHAALLAGLLSHVGMRDDQRDRKSRDRAR
MMAR_2947|M.marinum_M CRDLGLAESGPQSDDAAEPARVHAALLAGLLSHVGMRREDG---------
MAV_2737|M.avium_104 AGELGITE---ESGEPADPARVHAALLAGLLSHVGMRREDS---------
. ::*: * . : * * :**** ******:*:* :
MSMEG_6587|M.smegmatis_MC2_155 ----RDYTGARNTKFVLAPGSVLTKKPPRWIVVADLVETSRLFGRIAARI
MAB_0056c|M.abscessus_ATCC_199 ----RDYSGARNTKFVLAPGSVLTKKPPRWVVVADLVETSRLFGRIAARV
Mflv_1063|M.gilvum_PYR-GCK ----RAYDGARNAKFVLAPGSVLSRRPPRWVVVADLVETSRLFGRTAARI
Mvan_5766|M.vanbaalenii_PYR-1 ----RSYQGARNAKFVLAPGSVLSKRPPRWVVVADLVETSRLFGRTAARI
TH_0802|M.thermoresistible__bu DRGPREYLGARNTRFVLAPGSVLTRQPPRWVVVAELVETSRLYGRIAARI
MMAR_2947|M.marinum_M ----REYLGARNSRFVLAPGSALTKRPPRWVVVAELVETSRLYGRIAARI
MAV_2737|M.avium_104 ----REFAGARNSRFVLAPGSVLSKRPPRWVVVAELVETTRLYGRTAARI
* : ****::*******.*:::****:***:****:**:** ***:
MSMEG_6587|M.smegmatis_MC2_155 EPEAVERVAGHLVQRTYSEPHWDARRGAVMASERVTLYGLPLVPRRRVNY
MAB_0056c|M.abscessus_ATCC_199 EPQAVERLAGDLVHKTYSEPHWDAERGAVMAFERVTLYGLPLVPRRRVGY
Mflv_1063|M.gilvum_PYR-GCK EPDTVERVAGHVLQRTYSEPHWDAERGAVMAYERVTLYGLPLVARRRIGY
Mvan_5766|M.vanbaalenii_PYR-1 EPETVERVAGHLVQRTYSEPHWDAERGAVMAYERVTLYGLPLVARRRVGY
TH_0802|M.thermoresistible__bu DPQTVERVGAHLVQRGYSEPHWDAQRGAAMAYERVTLYGLPVVPRRRVGY
MMAR_2947|M.marinum_M QPEVVERVAGELAQRSHSEPHWDTKRGEVMAYERVTLYGLPLAARRRVGY
MAV_2737|M.avium_104 QPEAVERVAGDLVQRSHSEPHWDPDRGEVMAYERVTLYGLPLVSRRRVGY
:*:.***:...: :: :******. ** .** *********:..***:.*
MSMEG_6587|M.smegmatis_MC2_155 AQIDPVVSRELFIRHALVEGEWQTRHHFFRDNARLREEVEEMEERARRRD
MAB_0056c|M.abscessus_ATCC_199 AQVDPELARELFIRHALVEGDWQSKHHFLRDNARLLRELSELEDKARRRD
Mflv_1063|M.gilvum_PYR-GCK GQVDPEVSRDLFIRHALVEGDWRTKHHFFRDNARLREELAGLEDRARRRD
Mvan_5766|M.vanbaalenii_PYR-1 AQVDPEAARDLFIRHALVEGDWQTKHHFFRDNARLREELAELEDRARRRD
TH_0802|M.thermoresistible__bu ATVEPEVARELFIRHALVEGDWHTRHRFFHDNARLRDELAALEDRARRRD
MMAR_2947|M.marinum_M SRVEPTVARQLFIRHALVEGDWQTRHHFFADNARLRTQLEELEERARRRG
MAV_2737|M.avium_104 ARIDPVLARELFIRHALVEGDWHTRHRFFADNARLRGELEELEERARRRD
. ::* :*:**********:*:::*:*: ***** :: :*::****.
MSMEG_6587|M.smegmatis_MC2_155 LLVGDDEIFALYDARIPADVVSARHFDAWWKKQRHKTPDLLTFTREDLLR
MAB_0056c|M.abscessus_ATCC_199 LLVGDDEIYALYAARIPADAVSARHFDAWWRKQRHKTPNLLTFTRDELLR
Mflv_1063|M.gilvum_PYR-GCK LLVGDDEIFAFYDARIPADVVSSRHFDGWWRKERHRNPDLLTLTREDLLR
Mvan_5766|M.vanbaalenii_PYR-1 LLVGDDEVFAFYDARVPADVVSARHFDGWWRKQRHRTPGLLTMTRDDLLR
TH_0802|M.thermoresistible__bu LLVGDDEIYEFYAARIPDTVVSARHFDAWWKKQRHRTPDLLTMRREDLLR
MMAR_2947|M.marinum_M LLVDDDDVYELYDARIPAEVVSARHFDTWWKKQRHQTPDLLTFTRNELLR
MAV_2737|M.avium_104 LVVGDDDVYALYDARIPADVVSARHFDAWWKKQRHQTPDLLTFTRADLLR
*:*.**::: :* **:* .**:**** **:*:**:.*.***: * :***
MSMEG_6587|M.smegmatis_MC2_155 TDDA----ADHPDTWRAGDLELPLSYRYQPGAADDGVTVHVPVGVLARLG
MAB_0056c|M.abscessus_ATCC_199 TADAG---ADMPDTWRAGDVALPVTYRFEPGAADDGVSVHIPVEVLARLG
Mflv_1063|M.gilvum_PYR-GCK NVSDTHTDTEKPDTWQAGDLNLPLSYRFDPGSSDDGVTVHVPVDVLARLG
Mvan_5766|M.vanbaalenii_PYR-1 NEAG----AEQPDAWQAGDLSLPLSYRYDPGSADDGVTVHVPVDVLARLG
TH_0802|M.thermoresistible__bu TDDAD---AEQPDTWQSGDVELPLSYRFEPGAADDGVTVHVPVHVLARLG
MMAR_2947|M.marinum_M SDDPA--DTDRPDAWQTDDAALPLTYRFEPGAADDGVTVHVPVDVLARLG
MAV_2737|M.avium_104 TEETG--DADRPDSWRAGDVTLPLTYRFQPGAADDGVTVHVPIDVLARLG
. :: **:*::.* **::**::**::****:**:*: ******
MSMEG_6587|M.smegmatis_MC2_155 GENFAWQVPALREELVTALIKSLPKDLRRNFVPAPDTARAILPHLDPGTG
MAB_0056c|M.abscessus_ATCC_199 GDEFAWHVPALREELVTALIKSLPKDLRRNFVPAPDTARAVLQNLDPGSE
Mflv_1063|M.gilvum_PYR-GCK GDDFAWQVPALREELLTALIKSLPKDLRRNFVPAPDTARALLNAITPDSG
Mvan_5766|M.vanbaalenii_PYR-1 GDEFAWQVPALREELLTALIKSLPKDLRRNFVPAPDTARALLASITPDSG
TH_0802|M.thermoresistible__bu GDDFAWQVPALREELITELIRSLPKDLRRNFVPAPDTARALLTTLQPGDE
MMAR_2947|M.marinum_M DDQFAWQVPAFREELVTELIRSLPKDLRRNFVPAPDAARAVLAGIDPAKE
MAV_2737|M.avium_104 GDEFAWQVPALREELVTALIRSLPKDLRRNFVPAPDTARAVLGAIDPAAE
.::***:***:****:* **:***************:***:* : *
MSMEG_6587|M.smegmatis_MC2_155 SLLDAVQRELRRRTGILVPVDAFDLDKVPPHLRVTFAVENSDGAEVARGK
MAB_0056c|M.abscessus_ATCC_199 PLVYALARELHRLSGIRVPTDAFDLDKVPPHLRVTFVVESADGAEVARDK
Mflv_1063|M.gilvum_PYR-GCK PLLDAVQRELRRRTGILVPIDAFDLSKLPPHLRVTFAVEDADGSVVSRGK
Mvan_5766|M.vanbaalenii_PYR-1 PLLDAIQRELRRRTGILVPIDAFDLDKLPPHLRVTFAVEAADGTVVSRGK
TH_0802|M.thermoresistible__bu PLLEAVQRELHRRTGILVPIDAFDLDRLPAHLRVTFAVEGPDGTEVARGK
MMAR_2947|M.marinum_M PLLQALQRELRRRSGVLVPIDAFDLTKLPPHLRVNFAVESADGTEVARGK
MAV_2737|M.avium_104 PLLPALQRELRRRSGVSVPIDAFDLDKLPAHLRMTFAVESTDGTELARGK
.*: *: ***:* :*: ** ***** ::*.***:.*.** .**: ::*.*
MSMEG_6587|M.smegmatis_MC2_155 DIETLREQLAVPVAQAVAEAVDDGLQRTGLRAWPDDLDELPRTVENVSG-
MAB_0056c|M.abscessus_ATCC_199 NLSALQQRLADSTRQAVATAVAGQWERSGLRAWPGDLPELPHTVEQVSG-
Mflv_1063|M.gilvum_PYR-GCK NLDELQQQLAAPTRQAVAATVAGDLERTGLRDWPADLDELPRVVESAGAS
Mvan_5766|M.vanbaalenii_PYR-1 DLGELQQQLAAPSRQAVAATVAGDLERTGLRDWPPELDELPRVVERKGAT
TH_0802|M.thermoresistible__bu DLAALQERLAGTTRRAVADAVGADLERTGLRDWPEDLEELPRTVERR---
MMAR_2947|M.marinum_M DLKALQAELAAPAQRAVADAVAGDLERKGLRAWPDDVDELARVVERSVG-
MAV_2737|M.avium_104 DLRALQERLTTPARQAVADTVGARLQRTGLRGWPDDLDELPRVVQRSLD-
:: *: .*: . :*** :* :*.*** ** :: **.:.*:
MSMEG_6587|M.smegmatis_MC2_155 GHTVRGYPGFVDTGKAVDIRVFANPIERDAAMGPGLRRLLRLSVPSPVKA
MAB_0056c|M.abscessus_ATCC_199 GHTVRGYPALVDAGTSVDIHVFATESEQRAAMATGARRLLRLSIPSPMKT
Mflv_1063|M.gilvum_PYR-GCK EHVVRGYPALVPAGASVAIKVFATKDEQDAVMARGSRRLLLLAAPSVTKT
Mvan_5766|M.vanbaalenii_PYR-1 GHLVRGYPALVDAGKAADIKVFATKAEQDAAMGPGCRRLLLTAGPSVTKG
TH_0802|M.thermoresistible__bu --AVRGYPALVDTGPAVDLRVLPTEAEQSAAMRPGLRRLLRLTVPSPVKA
MMAR_2947|M.marinum_M GRAVRGFPGFVDQGPHVDLRVFATSAERDHAMAPGTRRLVRLGVAAPLKA
MAV_2737|M.avium_104 GRTVRGYPAFVDTGAAVDLRVFATSAEQVRMMGAGLRRLVRLSIPSPAKA
***:*.:* * . ::*:.. *: * * ***: .: *
MSMEG_6587|M.smegmatis_MC2_155 IERGLGTRTRLALNANPDGNLAALLEDCADAAADTLVRQIVWTRADFTVL
MAB_0056c|M.abscessus_ATCC_199 VERGLDPRSRLLLSSNPDGPLSALLDDCADAATDLLAPVPVWTRAEFTAL
Mflv_1063|M.gilvum_PYR-GCK VERGLDTRTRLLLGNNPDGSLSALLEDCAQAAVQILVPDPVWTKKEFAAA
Mvan_5766|M.vanbaalenii_PYR-1 VERSLDTRTRLVLGTNPDGSLAALIDDCAAAAVQVLVPAPAWTREEFAAA
TH_0802|M.thermoresistible__bu VERALDPRARLVLGNNPHGSPTALLEDCADAAVDVLAQTPVWTRAEFLAV
MMAR_2947|M.marinum_M VERQLDPRTRLLLGTNPDGSLTALLDDCADAATDALTPGPVWTRAEFVAL
MAV_2737|M.avium_104 IARQLDPRTRLALGGNPDGSLPALLEDCADAATDALVPQPVWTRDEFTAL
: * *..*:** *. **.* .**::*** **.: *. .**: :* .
MSMEG_6587|M.smegmatis_MC2_155 VQRAAKELGATTQAAVTRVEKVLAAAHEVGVALPDKPPAAQVDAVEDIRL
MAB_0056c|M.abscessus_ATCC_199 RDRAAQSLAATTLDIVRRVEKVVQAWSELQVALPTKAPAAQSEAIADMRD
Mflv_1063|M.gilvum_PYR-GCK RQRLAAGLAQTTADIVRRVQKVLAALHEVELAVPKQPSPAQADAIADIRA
Mvan_5766|M.vanbaalenii_PYR-1 RKRLAANIVATTADVVRRVEKVLVALHEVEVALPSKPTPAQADAIADIRG
TH_0802|M.thermoresistible__bu RDRVAAKLAATTQNVVGRVQHVLAAAQRVQLALPDPPPPTQADAVADIRD
MMAR_2947|M.marinum_M CADVAKALVPMTIDIVGRVAKVLAAAQEVQLALPAKPPSAQTDAIADIRA
MAV_2737|M.avium_104 RQRVAKTLGPNTIELVGRVEQVLAAAQQVQLALPATPPPAQADAVADIRA
* : * * ** :*: * .: :*:* ...:* :*: *:*
MSMEG_6587|M.smegmatis_MC2_155 QLERLLPKGFVTATGVARLTDLTRYVTAIARRLERLPHAIGADRERMARV
MAB_0056c|M.abscessus_ATCC_199 QLDRLLDAGFVAATGAARLTDLARYVNAIGKRLERLPQGVEADRERMHRV
Mflv_1063|M.gilvum_PYR-GCK QLSRLVPTGFVTATGAAKLGDLTRYLTAIDRRLERLPHALGADRERMDRV
Mvan_5766|M.vanbaalenii_PYR-1 QLARLVPPGFVAATGAARLGDLARYLTAIGRRLERLPHAPAADRERMERV
TH_0802|M.thermoresistible__bu QLGRLLPAGFVTASGAGRLADLTRYLTAIERRLERLPHAVAADRDRMARV
MMAR_2947|M.marinum_M QLDRLVPPGFVAATGRAHLGDLARYLIAIRRRLDRLPHAVAADRDRMERV
MAV_2737|M.avium_104 QLDRLLPAGFVTATGGARLGDLTRYLSAIRRRLDGLARAGQADRDRMRRV
** **: ***:*:* .:* **:**: ** :**: *.:. ***:** **
MSMEG_6587|M.smegmatis_MC2_155 HAVEDAYDDLVRALSPARAAAHDVSAIAWQIEELRVSLWAQQLGTPRPVS
MAB_0056c|M.abscessus_ATCC_199 HAVEDAYDDLLRDLPAGRADDPDIRDIAWFIEELRVSLWAQQLGTARAVS
Mflv_1063|M.gilvum_PYR-GCK AAVQDAYDDLRAALSPARAAAPEVVDIAWMIEELRVSLWAQQLGTARPIS
Mvan_5766|M.vanbaalenii_PYR-1 AAVQAEYDDLRRALSPGRAAATDVQDIARMIEELRVSLWAQQLGTARPVS
TH_0802|M.thermoresistible__bu HAVQEAYEELVAAVSPARAARPDIAEIRWMIEELRVSLWAQQLGTARPVS
MMAR_2947|M.marinum_M QAVVEAYDELVHALPLARASAADVRDIGRQIEELRVSLWAQQLGTPRPVS
MAV_2737|M.avium_104 HAAQHAYDELVDELPEAAKTAADVRDIAWQIEELRVSLWAQQLGTPRPVS
*. *::* :. . :: * ***************.*.:*
MSMEG_6587|M.smegmatis_MC2_155 EQRIYKAIDAVAR------------
MAB_0056c|M.abscessus_ATCC_199 EQRILKAINAAR-------------
Mflv_1063|M.gilvum_PYR-GCK EQRIYRALNAAETGR----------
Mvan_5766|M.vanbaalenii_PYR-1 EQRIYRAIDAVGVAGPGGGR-----
TH_0802|M.thermoresistible__bu EQRIYRAIDGVQL------------
MMAR_2947|M.marinum_M EQRIYRAIDAVHHSDTER-------
MAV_2737|M.avium_104 EQRIHRAIDAARQRYRVALPPTLNL
**** :*::..