For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SGRTLGRQIRDALPLLPFVAVVTVFLIVPTVTVIVMAFFADGSFSLERIFALFTGSALTALGKSVILSAS TALLGAVLGAALAWLIVSSPPASMLRRAVLALCSVLAQFGGVALAFAFLATIGLNGVLTLWVRQLFGWDL AGSGWLYGLAGLILVYTYFQIPLMVIVFIPALEGLREQWREAAVNLGASTWQYLREVAVPLLMPAFLGSA LLLFANAFAAYATAAALVSQGSPIVPLLIRAALTSEVVMGQSGFAYALALEMIVVVAVVMFAYNAMMRRT ARWLR*
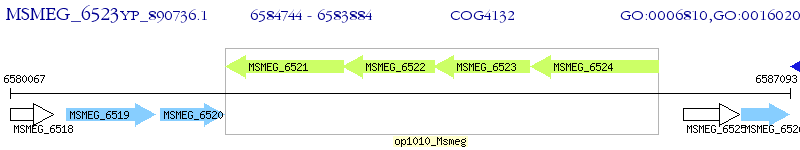
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6523 | - | - | 100% (286) | ABC transporter, membrane spanning protein |
| M. smegmatis MC2 155 | MSMEG_6669 | - | 5e-27 | 31.45% (283) | ABC transporter, permease protein |
| M. smegmatis MC2 155 | MSMEG_2015 | modB | 2e-14 | 26.71% (277) | molybdate ABC transporter, permease protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1889 | modB | 1e-11 | 26.24% (263) | molbdenum-transport integral membrane protein ABC MODB |
| M. gilvum PYR-GCK | Mflv_1081 | - | 1e-127 | 80.43% (281) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv1858 | modB | 1e-11 | 26.24% (263) | molbdenum-transport integral membrane protein ABC |
| M. leprae Br4923 | MLBr_01426 | - | 7e-09 | 22.18% (284) | putative ABC-transport protein, inner membrane component |
| M. abscessus ATCC 19977 | MAB_2434 | - | 6e-15 | 29.15% (271) | molybdenum ABC transporter permease |
| M. marinum M | MMAR_3717 | cysT | 2e-11 | 23.57% (280) | sulfate-transport integral membrane protein ABC transporter |
| M. avium 104 | MAV_1783 | cysT | 2e-10 | 23.05% (269) | sulfate ABC transporter, permease protein CysT |
| M. thermoresistible (build 8) | TH_3819 | - | 3e-11 | 25.68% (257) | PUTATIVE NifC-like ABC-type porter |
| M. ulcerans Agy99 | MUL_3660 | cysT | 2e-11 | 23.93% (280) | sulfate-transport integral membrane protein ABC transporter |
| M. vanbaalenii PYR-1 | Mvan_5741 | - | 1e-125 | 79.36% (281) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1081|M.gilvum_PYR-GCK --------------------------MS---LARRLRDSLPLLPFLIVVV
Mvan_5741|M.vanbaalenii_PYR-1 --------------------------MS---LARRLRDSLPLLPFLTVVV
MSMEG_6523|M.smegmatis_MC2_155 --------------------------MSGRTLGRQIRDALPLLPFVAVVT
Mb1889|M.bovis_AF2122/97 -----------------------------------MHPPTDLPRWVYLPA
Rv1858|M.tuberculosis_H37Rv -----------------------------------MHPPTDLPRWVYLPA
TH_3819|M.thermoresistible__bu -----------------------------------MN-ASTLPRWIYLPA
MAB_2434|M.abscessus_ATCC_1997 -----------------------------------MTPAPGLPRWVYVPA
MMAR_3717|M.marinum_M MTVTAHSGQSDPPAVAPDPGQRADGGPQWISGLGRSGRRGATALQVGVSM
MUL_3660|M.ulcerans_Agy99 MTVTAHSGQSDPPAVAPDLGQRADGGPQWISGPGRSGRRGATALQVGVSM
MAV_1783|M.avium_104 MTAITP----DPLAIRPELG--GDPGHQPVPGRGR----GTTALRVGVAT
MLBr_01426|M.leprae_Br4923 -MTSVETTAVPEPSIAKNHASLPPSRRRAWAGRMFIAPNLASVSVFMLFP
. :
Mflv_1081|M.gilvum_PYR-GCK IFLIIPTVTVVIGAVYSDGVFSLDRIAALFTG--TALSALADSVLLSGST
Mvan_5741|M.vanbaalenii_PYR-1 IFLIIPTATVVVGSVYTDGVFSLDRIEALFTG--TALSALLDSVLLSGAT
MSMEG_6523|M.smegmatis_MC2_155 VFLIVPTVTVIVMAFFADGSFSLERIFALFTG--SALTALGKSVILSAST
Mb1889|M.bovis_AF2122/97 IAGIVFVAMPLVAIAIRVD---WPRFWALITTP-SSQTALLLSVKTAAAS
Rv1858|M.tuberculosis_H37Rv IAGIVFVAMPLVAIAIRVD---WPRFWALITTP-SSQTALLLSVKTAAAS
TH_3819|M.thermoresistible__bu AVGATLLALPVFALAGRVD---WPRFWSLICSE-SSMTALALSLRTATAS
MAB_2434|M.abscessus_ATCC_1997 AIGAALIVLPLLALVVKAD---WGQFPALVTSP-ASQAALLLSVRTCLAS
MMAR_3717|M.marinum_M VWLSVIVLLPLAAIAWQAADGGWHAFWLSVSSH-AALESLRVTLTVSAGV
MUL_3660|M.ulcerans_Agy99 VWLSVIVLLPLAAIAWQAADGGWHAFWLSVSSH-AALESLRVTLTVSAGV
MAV_1783|M.avium_104 VWLSVIVLLPLAAIAWQSAGGGWHVFWLAVTSN-AALESFRVTLTISAGV
MLBr_01426|M.leprae_Br4923 LGFSLYMSFQKWDMFTPPVFVGLANFQNLFTSDPLFLIALCNSVVFTVGT
: . :: :: .
Mflv_1081|M.gilvum_PYR-GCK ALIGAFLGAVMAWLIISSPP-ASMLRRAVLSLCSVLAQFGGVALAFAFLA
Mvan_5741|M.vanbaalenii_PYR-1 ALLGAVFGAVMAWLIVSSPS-TSMVRRAVLSLCSVLAQFGGVALAFAFLA
MSMEG_6523|M.smegmatis_MC2_155 ALLGAVLGAALAWLIVSSPP-ASMLRRAVLALCSVLAQFGGVALAFAFLA
Mb1889|M.bovis_AF2122/97 TVLCVLLGVPMALVLARSR---GRLVRSLRPLILLPLVLPPVVGGIALLY
Rv1858|M.tuberculosis_H37Rv TVLCVLLGVPMALVLARSR---GRLVRSLRPLILLPLVLPPVVGGIALLY
TH_3819|M.thermoresistible__bu TVLCLVLGVPMALALARGG---SPILRWIRPLVLLPLVLPPVVGGIALLY
MAB_2434|M.abscessus_ATCC_1997 TALCLLFGVPLAVVLARGP---GRMTRLLRPMILLPLVLPPVVGGIALLY
MMAR_3717|M.marinum_M TLLNTVFGLLIAWVLIRDE---FVGKRLVDAVIDLPFALPTIVASLVMLA
MUL_3660|M.ulcerans_Agy99 TLLNAVFGLLIAWGLIRDE---FVGKRLVDAVIDLPFALPTIVASLVMLA
MAV_1783|M.avium_104 TVLNLVFGLVIAWVLVRDE---FPGKRLVDAIIDLPFALPTIVASLVMLA
MLBr_01426|M.leprae_Br4923 VIPTVLISLVVAGVLNQKVKGIGIFRTIVFLPLAISSVVMAVVWQFIFNT
. .:. :* : : : . :. : :
Mflv_1081|M.gilvum_PYR-GCK TVGLNGVLTLWVQQAFGFNLAGSGWLYSLAGLTLVYTYFQIPLMVIVFIP
Mvan_5741|M.vanbaalenii_PYR-1 TVGLNGVLTLWVQQVFGFNLAGSGWLYSLSGLILVYTYFQIPLMVIVFVP
MSMEG_6523|M.smegmatis_MC2_155 TIGLNGVLTLWVRQLFGWDLAGSGWLYGLAGLILVYTYFQIPLMVIVFIP
Mb1889|M.bovis_AF2122/97 AFGRLGLIGRYL------EAAGISIAFSTAAVVLAQTFVSLPYLVISLEG
Rv1858|M.tuberculosis_H37Rv AFGRLGLIGRYL------EAAGISIAFSTAAVVLAQTFVSLPYLVISLEG
TH_3819|M.thermoresistible__bu AFGRMGLIGGYL------EAVGIQIAFSTTAVVLAQTFVSLPFLVLALEG
MAB_2434|M.abscessus_ATCC_1997 AFGKLGLLGHYL------DAAGIRIAYSTSAVVLAQTFVSLPFLVISLQG
MMAR_3717|M.marinum_M LYGHN-------------SPLGLHLQHTVWGVGVALAFVTLPFVVRAVQP
MUL_3660|M.ulcerans_Agy99 LYGHN-------------SPLGLHLQHTVWGVGVALAFVTLPFVVRAVQP
MAV_1783|M.avium_104 LYGNN-------------SPVGLHLQHTAAGVAVALAFVTLPFVVRAVQP
MLBr_01426|M.leprae_Br4923 HNGLLNIMLGWIG--IGPIPWLVNPGWAMASLCIVSVWRSVPFATVVLLA
* .: :. .: :* . .
Mflv_1081|M.gilvum_PYR-GCK ALEGLRGQWREAAVSLGASTWAYWREVAIPLLTPAFLGSALLLFANAFAA
Mvan_5741|M.vanbaalenii_PYR-1 ALEGLREQWREAAVSLGASTWAYWREVAIPLLTPAFLGSALLLFANAFAA
MSMEG_6523|M.smegmatis_MC2_155 ALEGLREQWREAAVNLGASTWQYLREVAVPLLMPAFLGSALLLFANAFAA
Mb1889|M.bovis_AF2122/97 AARTAGADYEVVAATLGARPGTVWWRVTLPLLLPGVVSGSVLAFARSLGE
Rv1858|M.tuberculosis_H37Rv AARTAGADYEVVAATLGARPGTVWWRVTLPLLLPGVVSGSVLAFARSLGE
TH_3819|M.thermoresistible__bu AARTAGTGYEVVAATLGATPTTVWWRVTLPLLAPGLVSGAVLAFARALGE
MAB_2434|M.abscessus_ATCC_1997 ALTTTGQRYDEVAASLGAPPTTVLRRVSLPLVMPALASGTVLAFARSLGE
MMAR_3717|M.marinum_M VLLELDRETEEAAASLGATGWKVFTSVIFPSLMPALLSGAGLAFSRAIGE
MUL_3660|M.ulcerans_Agy99 VLLELDRETEEAAASLGATGWKVFTSVIFPSLMPALLSGAGLAFSRAIGE
MAV_1783|M.avium_104 VLLELDRETEEAAASLGASGPKIFTSVVLPALLPALLSGAGLAFSRAIGE
MLBr_01426|M.leprae_Br4923 AMQEVPKTVYEAAKIDGAGEIRQFISITVPFIRGAISFVVVISFIHAFQT
. .* ** : .* : .. : * .::
Mflv_1081|M.gilvum_PYR-GCK YATAAALVS--QGSPILPLLIRASLVSEVV-LGQAGFAYALALEMIVVVA
Mvan_5741|M.vanbaalenii_PYR-1 YATAAALVS--QGSPILPLLIRASLVSEVV-LGQAGFAYALALEMIVVVA
MSMEG_6523|M.smegmatis_MC2_155 YATAAALVS--QGSPIVPLLIRAALTSEVV-MGQSGFAYALALEMIVVVA
Mb1889|M.bovis_AF2122/97 FG-ATLTFA--GSRQGVTRTLPLEIYLQRV-TDP-DAAVALSLLLVVVAA
Rv1858|M.tuberculosis_H37Rv FG-ATLTFA--GSRQGVTRTLPLEIYLQRV-TDP-DAAVALSLLLVVVAA
TH_3819|M.thermoresistible__bu FG-ATLTFA--GSRQGVTRTLPLEIYLQRE-ADP-EAAVALSMVLIAVAA
MAB_2434|M.abscessus_ATCC_1997 FG-ATLTFA--GSRQGVTRTLPLEIYLQRE-SDP-QAAVALSLLLVAVAA
MMAR_3717|M.marinum_M FG-SVVLIG--GAVPGETEVSSQWIRTLIE-NDDRTGAAAISIVLLSISF
MUL_3660|M.ulcerans_Agy99 FG-SVVLIG--GAVPGETEVSSQWIRTLIE-NDDRTGAAAISIVLLSISF
MAV_1783|M.avium_104 FG-SVVLIG--GAVPGKTEVSSQWIRTLIE-NDDRTGAAAISIVLLAISF
MLBr_01426|M.leprae_Br4923 FDLVYVLNGPNGGPELATYVLGIMLFQHAFSFLEFGYASALALVTFAILL
: . . . : * *::: . :
Mflv_1081|M.gilvum_PYR-GCK VVMVAYNLLVRRSSRWLT----
Mvan_5741|M.vanbaalenii_PYR-1 VVMAAYNLLVRRSSRWLS----
MSMEG_6523|M.smegmatis_MC2_155 VVMFAYNAMMRRTARWLR----
Mb1889|M.bovis_AF2122/97 LVVLGVGART--PIGTDTR---
Rv1858|M.tuberculosis_H37Rv LVVLGVGART--PIGTDTR---
TH_3819|M.thermoresistible__bu VVVIGLGARR--LRAGGWS---
MAB_2434|M.abscessus_ATCC_1997 VVLAVVGARTRRPAGGGYE---
MMAR_3717|M.marinum_M AVLLVLRVLGARAAKREELAP-
MUL_3660|M.ulcerans_Agy99 AVLLVLRVLGARAAKREELAP-
MAV_1783|M.avium_104 VVLLILRVVGGRAAKREELAA-
MLBr_01426|M.leprae_Br4923 VLIVLQLLLNRRHSWEVSGGLS
::