For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TPSKILDLVAAAFGELLRRAGVATSPAEVIEVRRVLGLLGARDQEALRAALRATCAKYVREQRGFDRAFD AMFGVASIVTEHDVADPSRAHTGSGLPDHLEIDEDQEIGRYAEYNERAAEVGDHFDTPDADKGFNPHKDD DDFSVSGADNQLSVSAESDAGRRGVRYTVEVDRAASSLAGDLADSVAPVVAGALHWDDPESILAWLDAYD PARTYGDDAGDQPLTAEQLDQLTQAVESFVAALADRCGLTAEPDTPAGPTDATVATDVDLACHEVLRRMR GAPRPRPAEMRGRLDMRRTTRHSLRTDGVPFRLITRTPRPDRVRLLVIADVSLSVRPVTAFTLRLAQAMH RRAHRCRVMAFVDRPVDVTEILLQTGGDDALAAVLAEAQIDLEASSDYGQVFDEMLSAQAEALDSRTAVL IVGDGRSPGLPPRTDSLRALRRRVHRLAWITPEPTRYWRQATCAMPDYAEVCDKVVVARDAEQLIARAGE LAHALS*
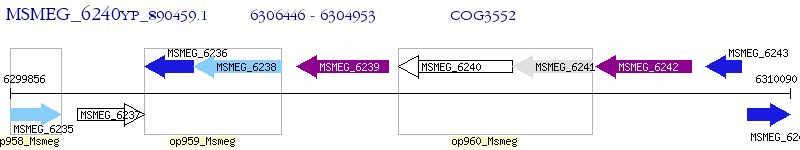
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6240 | - | - | 100% (497) | hypothetical protein MSMEG_6240 |
| M. smegmatis MC2 155 | MSMEG_4582 | - | 3e-31 | 26.88% (491) | hypothetical protein MSMEG_4582 |
| M. smegmatis MC2 155 | MSMEG_0751 | - | 5e-08 | 26.00% (250) | hypothetical protein MSMEG_0751 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2449c | - | 4e-32 | 27.47% (495) | hypothetical protein Mb2449c |
| M. gilvum PYR-GCK | Mflv_2646 | - | 5e-32 | 26.64% (488) | VWA containing CoxE family protein |
| M. tuberculosis H37Rv | Rv2425c | - | 4e-32 | 27.47% (495) | hypothetical protein Rv2425c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1620 | - | 9e-31 | 26.18% (489) | hypothetical protein MAB_1620 |
| M. marinum M | MMAR_3751 | - | 8e-32 | 27.59% (493) | hypothetical protein MMAR_3751 |
| M. avium 104 | MAV_1749 | - | 4e-30 | 27.80% (500) | hypothetical protein MAV_1749 |
| M. thermoresistible (build 8) | TH_0790 | - | 3e-31 | 27.31% (498) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3694 | - | 8e-33 | 27.79% (493) | hypothetical protein MUL_3694 |
| M. vanbaalenii PYR-1 | Mvan_5477 | - | 1e-53 | 30.88% (502) | VWA containing CoxE family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2449c|M.bovis_AF2122/97 MAARR-IRAARPLAPHGLPGHLVGFVEALRGSGISVGPSETVDAGRVMAT
Rv2425c|M.tuberculosis_H37Rv MAARR-IRAARPLAPHGLPGHLVGFVEALRGSGISVGPSETVDAGRVMAT
MMAR_3751|M.marinum_M MATRR-IRPSYPLAPHGLPGHLVGFVEALRGSGISVGPSETVDAGRVMAA
MUL_3694|M.ulcerans_Agy99 MATRR-IRPSYPLAPHGLPGHLVGFVEALRGSGISVGPSETVDAGRVMAA
MAV_1749|M.avium_104 MATRR-IRPSRPLAPHGLPGHLVGFVEALRGAGISVGPSETVDAGRVMAT
Mflv_2646|M.gilvum_PYR-GCK MPPRR-TRPPQPPAPHGIPGHLVEFVEALRKQGIAVGPSETVDAGQVMAT
TH_0790|M.thermoresistible__bu MAVRR-TRPPQPLAPHGIPGHLVEFIEALRAQGIYVGPSETVDAGRVVSV
MAB_1620|M.abscessus_ATCC_1997 MPPRKPPKAVLPLAPHGLPGHLVGFVEALREQGISVGPSETVDAGRVMTV
Mvan_5477|M.vanbaalenii_PYR-1 -----------------MEATLHRFVRLLRLGGVRVSIPEALDAMRCAGQ
MSMEG_6240|M.smegmatis_MC2_155 -----------MTPSKILDLVAAAFGELLRRAGVATSPAEVIEVRRVLGL
: * . ** *: .. .*.::. :
Mb2449c|M.bovis_AF2122/97 LG-LGDREVLREGIACAVLRRPDHRDTYDAMFDLWFPAA-----------
Rv2425c|M.tuberculosis_H37Rv LG-LGDREVLREGIACAVLRRPDHRDTYDAMFDLWFPAA-----------
MMAR_3751|M.marinum_M LG-FEDREVLREGIACAVLRRPDHRDTYDAMFDLWFPAA-----------
MUL_3694|M.ulcerans_Agy99 LG-FEDREVLREGIACAVLRRPDHRDTYDAMFDLWFPAA-----------
MAV_1749|M.avium_104 LG-LGNREVLREGLACAVLRRPDHRETYDAMFDLWFPAA-----------
Mflv_2646|M.gilvum_PYR-GCK LG-LSDRNVLREGLACAVLRRSDHRETYDALFDLYFPAA-----------
TH_0790|M.thermoresistible__bu LG-LGDREVLREGIACAVLRRPDHRDTYDALFDLYFPAA-----------
MAB_1620|M.abscessus_ATCC_1997 LG-LQSRSELREGLACAVLRRADHRPTFDVLFDLWFPPA-----------
Mvan_5477|M.vanbaalenii_PYR-1 PGVLSSRAVLRTALRVALVKDQRDEPIFDEIFEAFFSLVRVGGDGHDAHA
MSMEG_6240|M.smegmatis_MC2_155 LG-ARDQEALRAALRATCAKYVREQRGFDRAFDAMFGVASIVTEHDVADP
* .: ** .: : : .. :* *: * .
Mb2449c|M.bovis_AF2122/97 ----------LGARAVITTEDESAGSG--GLPPDDVEAMRQLLLDLLANN
Rv2425c|M.tuberculosis_H37Rv ----------LGARAVITTEDESAGSG--GLPPDDVEAMRQLLLDLLANN
MMAR_3751|M.marinum_M ----------LGARAVIT-EEQAGQSE--GLPPDDVEAMRQLLLDLLTDN
MUL_3694|M.ulcerans_Agy99 ----------LGARAVIT-EEQAGQSE--GLPPDDVEAMRQMLLDLLTDN
MAV_1749|M.avium_104 ----------LGARAVVTDEDAGPGSGEDALPPDDVEAMREMLLDLLTEN
Mflv_2646|M.gilvum_PYR-GCK ----------LGAKTVLDDD---EDGE--GLPPEDIEALRSALVDLLADN
TH_0790|M.thermoresistible__bu ----------LGARTVLDEDGEPTDDP--GLPAEDIEAMRAALLDLLADN
MAB_1620|M.abscessus_ATCC_1997 ----------LGARFVEVGEE--------GIDIDDIEQVRDELVDLLASD
Mvan_5477|M.vanbaalenii_PYR-1 HSHAHDDLVDTGELESFTLSEEPSETPEPGHEHGKPASIRDYFKQEDLAQ
MSMEG_6240|M.smegmatis_MC2_155 SRAHTG----SGLPDHLEIDEDQEIGR-YAEYNERAAEVGDHFDTPDADK
* . . : : .
Mb2449c|M.bovis_AF2122/97 QDLAGKDERLVEMIARIVEAYGKYSS---SRGPSFSSYQALKAMALD---
Rv2425c|M.tuberculosis_H37Rv QDLAGKDERLVEMIARIVEAYGKYSS---SRGPSFSSYQALKAMALD---
MMAR_3751|M.marinum_M PDLADMDQRLVGMIARIVETYGKYNS---SRGPSFSSYQALKAMALD---
MUL_3694|M.ulcerans_Agy99 PDLADMDQRLVGMIARIVETYGKYNS---SQGPSFSSYQALKAMALD---
MAV_1749|M.avium_104 PDLADMDERLVAMIARIVEAYGKYSS---SRGPSFSSYQALKAMALD---
Mflv_2646|M.gilvum_PYR-GCK EELANIDERLAAMIAQIVEAYGRYNS---SRGPSYSSYQALKAMSLD---
TH_0790|M.thermoresistible__bu EELANLDERLAMMIAKIVEAYGRYNS---SRGPSYSSYQALKAMALD---
MAB_1620|M.abscessus_ATCC_1997 ---AG-DLPLNSLIAQIVEAYGKYNS---TRGTSYSAYQAMKSLSLD---
Mvan_5477|M.vanbaalenii_PYR-1 RYNLHQEANKIDLAALTDEIAFSKDS---RNGADDSYRIQLSTDRLGGAG
MSMEG_6240|M.smegmatis_MC2_155 GFNPHKDDDDFSVSGADNQLSVSAESDAGRRGVRYTVEVDRAASSLAG--
: : . : .* .* : : *
Mb2449c|M.bovis_AF2122/97 ---------------ELEGKLLAGLLAPYGDEP--TATQEQIAKALAAQK
Rv2425c|M.tuberculosis_H37Rv ---------------ELEGKLLAGLLAPYGDEP--TATQEQIAKALAAQK
MMAR_3751|M.marinum_M ---------------ELEGKLLAGLLAPYGDEP--TPTQEQIAKAIAAQR
MUL_3694|M.ulcerans_Agy99 ---------------ELEGKLLAGLLAPYGDEL--TPTQEQIAKAIAAQR
MAV_1749|M.avium_104 ---------------ELEGKLLAGLLAPYGDEP--TPTQEEIAKALAAQR
Mflv_2646|M.gilvum_PYR-GCK ---------------DLEGRLLAGLLAPYGDEP--TPTQEQIAKAIAAQR
TH_0790|M.thermoresistible__bu ---------------DLEGRLLAGLLAPYGDEP--TPSQEQIAKAIAAQR
MAB_1620|M.abscessus_ATCC_1997 ---------------QIEAKLLTGLLGGGDEDSPVSPTDQAVAKAIAKER
Mvan_5477|M.vanbaalenii_PYR-1 TAGDLATAAGTPVDAELTIAEQEALLGWLDDET--DYGGDEFDAAELRKR
MSMEG_6240|M.smegmatis_MC2_155 ------------DLADSVAPVVAGALHWDDPES-ILAWLDAYDPARTYGD
: . * . : : *
Mb2449c|M.bovis_AF2122/97 IAQLRRMVDAETKRRTAEQLGREHVQMYGIPQLSENVEFLRASGEQLRQM
Rv2425c|M.tuberculosis_H37Rv IAQLRRMVDAETKRRTAEQLGREHVQMYGIPQLSENVEFLRASGEQLRQM
MMAR_3751|M.marinum_M VSQLRKMVDAETKRRTAEQLGRDHVQMYGIPQLSENVEFLRASGEQLRQM
MUL_3694|M.ulcerans_Agy99 VSQLRKMVDAETKRRTAEQLGRDHVQMYGIPQLSENVEFLRASGEQLRQM
MAV_1749|M.avium_104 ITQLRKLVDAETKRRTAEQLGRDHVQMYGIPQLSENVEFLRASGEQLRQM
Mflv_2646|M.gilvum_PYR-GCK IAQLRKMVEGETKRRTAEQLGRDHVQMYGVPQLSENVEFLRASGEQLRQM
TH_0790|M.thermoresistible__bu ILQLKQMVEAETKRRTAEQLGREHVQMYGVPQLAENVEFLRASGEQLRQM
MAB_1620|M.abscessus_ATCC_1997 ISKVRQLVEAETKRRTAEQLGRERVTAYGVPQLAENVEFLRASGEQLRNM
Mvan_5477|M.vanbaalenii_PYR-1 LAGVLENLPQALKRHLEALLALETKIVEGREQREARIDRIGE--RERAEL
MSMEG_6240|M.smegmatis_MC2_155 DAGDQPLTAEQLDQLTQAVESFVAALADRCGLTAEPDTPAGPTDATVATD
.: .
Mb2449c|M.bovis_AF2122/97 RRVVAPLARTLATRLAARRRRARAGSIDLRKTLRKSMSTGGVPIDLVLHK
Rv2425c|M.tuberculosis_H37Rv RRVVAPLARTLATRLAARRRRARAGSIDLRKTLRKSMSTGGVPIDLVLHK
MMAR_3751|M.marinum_M RRVVAPLARTLATRLAARRRRARAGSIDLRKTLRKSMSTGGVPIDVVLRK
MUL_3694|M.ulcerans_Agy99 RRVVAPLARTLATRLAARRRRARAGSIDLRKTLRKSMSTGGVPIDVVLRK
MAV_1749|M.avium_104 QRVVAPLARTLATRLAARRRRSRAGTIDLRKTLRKSMSTGGVPIDVVLAK
Mflv_2646|M.gilvum_PYR-GCK RRVVGPLARTLATRLAARRRRSRAGEIDLRKTLRKSMSTGGVPIDVVLKK
TH_0790|M.thermoresistible__bu QRVVAPLARMLATRLAARRRRARAGEIDLRKTLRRSMSTGGVPIDLVLKK
MAB_1620|M.abscessus_ATCC_1997 RRVVHPLARMLASRLAARRRRAHTGQIDLRRTLRKSMSTGGVPIDVVQKK
Mvan_5477|M.vanbaalenii_PYR-1 EDSLRRLARTLHGALTHRRRTAAAGRVDSGQTMRRNMRFDGVPFMPVTVR
MSMEG_6240|M.smegmatis_MC2_155 VDLACHEVLRRMRGAPRPRPAEMRGRLDMRRTTRHSLRTDGVPFRLITRT
. . * * :* :* *:.: .***: :
Mb2449c|M.bovis_AF2122/97 PRPARPELVVLCDVSGSVAGFSHFTLLLVHALRQQFSRVRVFAFIDSTDE
Rv2425c|M.tuberculosis_H37Rv PRPARPELVVLCDVSGSVAGFSHFTLLLVHALRQQFSRVRVFAFIDSTDE
MMAR_3751|M.marinum_M PRPARPELVVLCDVSGSVAGFSHFTLLLVHALRQQFSRVRVFAFIDTTDE
MUL_3694|M.ulcerans_Agy99 PRPARPELVVLCDVSGSVAGFSHFTLLLVHALRRQFSRVRVFAFIDTTDE
MAV_1749|M.avium_104 PRPARPELVVLCDVSGSVAGFSHFTLLLVHALRQQFSRVRVFAFIDTTDE
Mflv_2646|M.gilvum_PYR-GCK PHPARPELVVLCDVSGSVAGFSHFTLLLVHALRQQFSRVRVFAFIDTTDE
TH_0790|M.thermoresistible__bu PHPARPELAVLCDVSGSVAGFSHFTLMLVHALRQQFSRVRVFAFIDTTDE
MAB_1620|M.abscessus_ATCC_1997 PRPARPELVVLCDVSGSVAGFSHFTLLLVNALRQQFSRVRVFAFIDTTDE
Mvan_5477|M.vanbaalenii_PYR-1 RAEDRPRLVVLADVSLSVRATSRFTLNLVHGLQDLFTQVRSFVFVADVAE
MSMEG_6240|M.smegmatis_MC2_155 PRPDRVRLLVIADVSLSVRPVTAFTLRLAQAMHRRAHRCRVMAFVDRPVD
* .* *:.*** ** : *** *.:.:: : * :.*: :
Mb2449c|M.bovis_AF2122/97 VTHMFGPESDLAIAIQRITREAGVYARDGHSDYGNAFVSFMQGFPNVLSP
Rv2425c|M.tuberculosis_H37Rv VTHMFGPESDLAIAIQRITREAGVYARDGHSDYGNAFVSFMQGFPNVLSP
MMAR_3751|M.marinum_M VTHMFGPDADLAVAIQRITREAGVYSRDGHSDYGNAFVSFMQGFPNVLSP
MUL_3694|M.ulcerans_Agy99 VTHMFGPDADLAVAIQRITREAGVYSRDGHSDYGNAFVSFMQGFPNVLSP
MAV_1749|M.avium_104 VTHMFGPEADLAVAIQRITREAGVYSRDGHSDYGNAFASFEQKFPNVLSP
Mflv_2646|M.gilvum_PYR-GCK VTELFGPDSDLAVAVQRITREAAVFTRDGHSDYGHAFVSFLDKFPNVLSP
TH_0790|M.thermoresistible__bu VTDLFGPDADLAVAVQRITREAEVFTRDGHSDYGHAFVSFLDKYPNALSP
MAB_1620|M.abscessus_ATCC_1997 VTHLFTPDTDLAEAIVRITREAQVFTGDGHSDYGHAFKTFVEKFPTVLSP
Mvan_5477|M.vanbaalenii_PYR-1 TTELFRDHP--SEKALGLIFGGDVVDLAANSDYGTVFGEFVAEHSSALTR
MSMEG_6240|M.smegmatis_MC2_155 VTEILLQTG--GDDALAAVLAEAQIDLEASSDYGQVFDEMLSAQAEALDS
.*.:: . . **** .* : . .*
Mb2449c|M.bovis_AF2122/97 RSSLLVLGDGRTNYRNPATDVLADMVTASRHAHWLNPEPKHLWGSGDSAV
Rv2425c|M.tuberculosis_H37Rv RSSLLVLGDGRTNYRNPATDVLADMVTASRHAHWLNPEPKHLWGSGDSAV
MMAR_3751|M.marinum_M RSSLLVLGDGRTNYRNPATDVLADMVTASRHAHWLNPEPQHLWGSGDSAV
MUL_3694|M.ulcerans_Agy99 RSSLLVLGDGRTNYRNPATDVLADMVTASRHAHWLNPEPQHLWGSGDSAV
MAV_1749|M.avium_104 RTSLLVLGDGRTNYRNPEVELLSHMVTASRHAHWLNPEPKHLWGSGDSAV
Mflv_2646|M.gilvum_PYR-GCK RSSLLVLGDGRNNYRNPETDLLAHMVNAARHAHWLNPEPKHLWGSGDSAV
TH_0790|M.thermoresistible__bu RSSLLILGDGRNNYRNPELELLERMVDASRHAHWLNPEPRHLWGSGDSAV
MAB_1620|M.abscessus_ATCC_1997 RSALLILGDGRTNYRNPAVEVLSSMVSSSRHAHWLNPEPKNHWGTGDSAA
Mvan_5477|M.vanbaalenii_PYR-1 RTTVLVLGDGRTNAKDPNLAAFEEITRRARETVWLTPEPRYSWGLGSCEL
MSMEG_6240|M.smegmatis_MC2_155 RTAVLIVGDGRSPGLPPRTDSLRALRRRVHRLAWITPEPTRYWRQATCAM
*:::*::****. * : : :. *:.*** * . .
Mb2449c|M.bovis_AF2122/97 PRYQEVIT-MHECRSAKQLATVIDQLLPV----
Rv2425c|M.tuberculosis_H37Rv PRYQEVIT-MHECRSAKQLATVIDQLLPV----
MMAR_3751|M.marinum_M PRYQEVIA-MHECRSAKQLATVIDELLPV----
MUL_3694|M.ulcerans_Agy99 PRYQEVIA-MHECRSAKQLATVIDELLPV----
MAV_1749|M.avium_104 PRYQQVIP-MHECRSAKQLAAVIDQLLPV----
Mflv_2646|M.gilvum_PYR-GCK PRYQDVIT-MHECRSAKQLASVIDALLPV----
TH_0790|M.thermoresistible__bu PRYSEVIT-MHECRSAKQLAAVIDQLLPV----
MAB_1620|M.abscessus_ATCC_1997 KRYQDAIT-MHECRSAKQLAAVIDNLLPV----
Mvan_5477|M.vanbaalenii_PYR-1 PAYAEFCDRVRVVRDLTGLESAAHDFAAEAVGR
MSMEG_6240|M.smegmatis_MC2_155 PDYAEVCDKVVVARDAEQLIARAGELAHALS--
* : : *. * : :