For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSDRGLEFGRELLNCAVEGTPGDDNPLRHVADLAPRTGKPVPWPAWAHPDVVRALHDHGITAPWSHQLSA AQLAHDGRHVVISTGTASGKSLAYQLPILSALADDPRARVLYLSPTKALGHDQLRAAHALTESVAALRDV APAPYDGDSATEVRRFARERSRWIFSNPDMIHLSLLRNHSRWAVFLRNLRYIVVDECHYYRGIFGSNVAM VLRRLLRLCERYSANGATPTVIFASATTSSPAETAAELIGQTVVEVTEDGSPQGARTIALWEPALLPDLA GENGAPVRRSAGSEAGRVMADLVAEGARTLTFVRSRRGAELTALGARARLSETAPDLVEQVASYRAGYLA EDRRELEHALSDGRLRGLATTNALELGIDIAGLDAVVMAGFPGTVASFWQQAGRSGRRGQGALVVLIARD DPLDTYLVHHPAALLDKPIERVVIDPTNPHVLGPQLLCAAAELPLTEAEVRRWDAEAVAQRLVDDGLLRK RPAGYFPAPGVDPHPAVDIRGSTGGQIAILEADTGRMLGSAGAGQAASSVHPGAVYLHQGESYVVDSLDF EDGIAFVHAEDPGYTTFAREITDISVTGPGERVDHGPVTIGLVPVSVTNTVVGYLRRRMDGEVIDFVELD MPPRTLDTMAVMCTITPEALQDNGIEQLSVPGALHAAEHASIGLLPLVASCDRGDIGGVSTAVGPVDGLP SIFVYDGYPGGAGFADRGYRQLQTWWGATAAAIEACECPSGCPSCVQSPKCGNGNDPLDKAGAVKVLRLV LGALTK
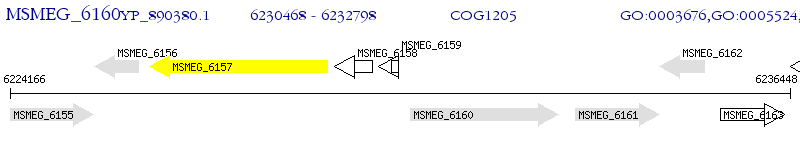
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6160 | - | - | 100% (776) | ATP-dependent rna helicase, dead/deah box family protein |
| M. smegmatis MC2 155 | MSMEG_1757 | - | 1e-20 | 29.08% (392) | DEAD/DEAH box helicase |
| M. smegmatis MC2 155 | MSMEG_1254 | - | 3e-14 | 28.11% (338) | DEAD/DEAH box helicase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3673 | - | 0.0 | 77.11% (769) | putative helicase |
| M. gilvum PYR-GCK | Mflv_1391 | - | 0.0 | 84.68% (770) | DEAD/DEAH box helicase domain-containing protein |
| M. tuberculosis H37Rv | Rv3649 | - | 0.0 | 76.98% (769) | helicase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0486c | - | 0.0 | 70.93% (767) | DEAD-box ATP dependent DNA helicase |
| M. marinum M | MMAR_5141 | - | 0.0 | 78.33% (766) | DEAD/DEAH box helicase |
| M. avium 104 | MAV_0515 | - | 0.0 | 78.11% (772) | ATP-dependent rna helicase, dead/deah box family protein |
| M. thermoresistible (build 8) | TH_0854 | - | 0.0 | 81.57% (776) | PROBABLE HELICASE |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5399 | - | 0.0 | 82.52% (778) | DEAD/DEAH box helicase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3673|M.bovis_AF2122/97 ----MASFGSHLLAAAVAGTPPGERPLRHVAELPPQAGRPRGWPEWAEPD
Rv3649|M.tuberculosis_H37Rv ----MASFGSHLLAAAVAGTPPGERPLRHVAELPPQAGRPRGWPEWAEPD
MAV_0515|M.avium_104 ----MASFGSELLAAALAGTTADEQPLRHVAELPPRSGRPHDWPAWAEPE
MMAR_5141|M.marinum_M ----MISFGRELLAVALAGTPTGEHPLHHVAELPPRAGQPSAWPQWAEPD
Mflv_1391|M.gilvum_PYR-GCK MTEPVPEFGRELLACAVEGTAADEHPLRHVADLPPRRATSHPWPPWADPD
Mvan_5399|M.vanbaalenii_PYR-1 MTDPGPDFGRELLACAVDGTAAGEYPLRHVADLPPRRARAQAWPHWADPD
MSMEG_6160|M.smegmatis_MC2_155 MSDRGLEFGRELLNCAVEGTPGDDNPLRHVADLAPRTGKPVPWPAWAHPD
TH_0854|M.thermoresistible__bu VEETGPNFGRELLACAVAGTAPDERPVRHIADVPPRRGRTRPWPSWADPD
MAB_0486c|M.abscessus_ATCC_199 ---MTVGYGRELLDLIVAGTPSGETPVRHICDIPARNAEVTSWPQWAHPD
:* .** : **. .: *::*:.::..: . ** **.*:
Mb3673|M.bovis_AF2122/97 VVDAFADRGISSPWSHQAEAAELAYAGRHVVIGTGPASGKSLAYQLPVLN
Rv3649|M.tuberculosis_H37Rv VVDAFADRGISSPWSHQAEAAELAYAGRHVVIGTGPASGKSLAYQLLVLN
MAV_0515|M.avium_104 VVGAFAARGIKSPWSHQFTAADLAWSGRHVVVSTGTASGKSLAYQLPALN
MMAR_5141|M.marinum_M VVRTFTQRGITAPWSHQSTAAELAHDGRHVVVSTGTASGKSLAYQLPVLA
Mflv_1391|M.gilvum_PYR-GCK VVQAFHDQGIRSLWSHQLAAADLAHDGRHVVLSTGTASGKSLAYQLPIAT
Mvan_5399|M.vanbaalenii_PYR-1 VVRAFHDRGVEALWSHQLAAAELARDGRHVVLSTGTASGKSLAYQLPILT
MSMEG_6160|M.smegmatis_MC2_155 VVRALHDHGITAPWSHQLSAAQLAHDGRHVVISTGTASGKSLAYQLPILS
TH_0854|M.thermoresistible__bu VVRVFGDRGITEPWEHQAVAADLAHSGRHVVLSTGTASGKSLAYQLPILS
MAB_0486c|M.abscessus_ATCC_199 AIAAFAAQGVTAPWSHQVAAATLAFAGRHVVVATGTASGKSVAYQLPILT
.: .: :*: *.** ** ** *****:.**.*****:****
Mb3673|M.bovis_AF2122/97 ALATDSRARALYLSPTKALGHDQLRAAHALAAAVPRLAD----VAPTAYD
Rv3649|M.tuberculosis_H37Rv ALATDSRARALYLSPTKALGHDQLRAAHALAAAVPRLAD----VAPTAYD
MAV_0515|M.avium_104 ALATDPRARVLYLSPTKALGHDQLRAAHALTAAIPRLREPGFAVAPTAYD
MMAR_5141|M.marinum_M ALAADPRARVLYLSPTKALGHDQLRTAQALVANVERLRD----VAPTAYD
Mflv_1391|M.gilvum_PYR-GCK ELKRNPRSRVLYLSPTKALGHDQLRAAAALTASVPGLDD----VAPTPYD
Mvan_5399|M.vanbaalenii_PYR-1 ALKENPRSRALYLSPTKALGHDQLRAAAALTAAVPGLAD----VAPTPYD
MSMEG_6160|M.smegmatis_MC2_155 ALADDPRARVLYLSPTKALGHDQLRAAHALTESVAALRD----VAPAPYD
TH_0854|M.thermoresistible__bu ALAADPRARVLYLSPTKALGHDQLRAAQELTGAVERLAT----VAPTAYD
MAB_0486c|M.abscessus_ATCC_199 ALASEPRARALYLAPTKALGHDQLRAVRELITGVDNFER----VQPCTYD
* :.*:*.***:***********:. * : : * * .**
Mb3673|M.bovis_AF2122/97 GDSPDEVRRFARERSRWLFSNPEMTHLSVLRNHARWAVLLRNLRFVIVDE
Rv3649|M.tuberculosis_H37Rv GDSPDEVRRFARERSRWLFSNPEMTHLSVLRNHARWAVLLRNLRFVIVDE
MAV_0515|M.avium_104 GDSPTEVRRFARERSRWVFSNPDMIHLSTLRNHARWAVLLRGLRFVIVDE
MMAR_5141|M.marinum_M GDSSDEVRRFARERSRWLFSNPDMIHLSILRNHARWAVLLRNLRYLIVDE
Mflv_1391|M.gilvum_PYR-GCK GDSPSEVRRFARERSRWIFSNPDMIHLSLLRNHARWAVFLRNLRYVVVDE
Mvan_5399|M.vanbaalenii_PYR-1 GDSPTEVRRFARERSRWIFSNPDMIHLSMLRNHARWAVFLRHLQYLVIDE
MSMEG_6160|M.smegmatis_MC2_155 GDSATEVRRFARERSRWIFSNPDMIHLSLLRNHSRWAVFLRNLRYIVVDE
TH_0854|M.thermoresistible__bu GDSPTEVRRFARERSRWIFSNPDMVHLAMLRNHGRWAVFLRHLRYVVVDE
MAB_0486c|M.abscessus_ATCC_199 GDSSNELRRFARESSRWIFTNPDMIHVSLLHNHTRWSAFLRGLRYLVVDE
***. *:****** ***:*:**:* *:: *:** **:.:** *:::::**
Mb3673|M.bovis_AF2122/97 CHYYRGVFGSNVAMVLRRLLRLCARYSAHP------TVIFASATTASPGA
Rv3649|M.tuberculosis_H37Rv CHYYRGVFGSNVAMVLRRLLRLCARYSAHP------TVIFASATTASPGA
MAV_0515|M.avium_104 CHYYRGVFGSHVAMVLRRLLRLCARYASAP------AVIFASATTDSPGA
MMAR_5141|M.marinum_M CHYYRGVFGSNVAMVLRRLLRLCARYSAHP------TVIFASATTDSPGA
Mflv_1391|M.gilvum_PYR-GCK CHYYRGIFGSNVAMVLRRLLRLCARYAPAG-DVSGPTVVFASATTAQPAI
Mvan_5399|M.vanbaalenii_PYR-1 CHYYRGIFGSNVAMVLRRLLRLCARYSATGADFSGPTVIFASATTAQPAA
MSMEG_6160|M.smegmatis_MC2_155 CHYYRGIFGSNVAMVLRRLLRLCERYSANG---ATPTVIFASATTSSPAE
TH_0854|M.thermoresistible__bu CHYYRGIFGSNVAMVLRRLLRLCQRYSAGG----SPTVFFASATTAAPAD
MAB_0486c|M.abscessus_ATCC_199 CHYYRGVFGSNVALILQRLLRLCERYGSHP------TVVFASATTANPGA
******:***:**::*:****** **.. :*.****** *.
Mb3673|M.bovis_AF2122/97 TAADLIGQPVVEVTEDGSPRGARTVALWEPALRSDVIGEHGAPVRRSAGA
Rv3649|M.tuberculosis_H37Rv TAADLIGQPVVEVTEDGSPRGARTVALWEPALRSDVIGEHGAPVRRSAGA
MAV_0515|M.avium_104 TAAELIGLPVREVTEDGSPQGARTVALWEPALRADLVGENGAPVRRSAGG
MMAR_5141|M.marinum_M TAAELIGQPVAEVTEDGSPQGARTVALWEPALRTDLTGENGAPVRRSAGT
Mflv_1391|M.gilvum_PYR-GCK TASELIGQTVAEVTEDGSPQGARTIALWEPALIPDLLGENGAPVRRSAGA
Mvan_5399|M.vanbaalenii_PYR-1 TASELIGQTVAEVTEDGSPQGARTVALWEPALIADLVGENGAPVRRSAGA
MSMEG_6160|M.smegmatis_MC2_155 TAAELIGQTVVEVTEDGSPQGARTIALWEPALLPDLAGENGAPVRRSAGS
TH_0854|M.thermoresistible__bu TASELIGQSVTEVTDDASPHGARTIALWEPPLLPDLVGENGAPVRRPAGA
MAB_0486c|M.abscessus_ATCC_199 AAQALIGQPVEEVVADGSPHGMRTVALWEPPLLP-ITGENGAPVRRPVST
:* *** .* **. *.**:* **:*****.* . : **:******...
Mb3673|M.bovis_AF2122/97 EAARVMADLIVEGAQTLTFVRSRRAAELTALGARARLVDIAPELSDTVAS
Rv3649|M.tuberculosis_H37Rv EAARVMADLIVEGAQTLTFVRSRRAAELTALGARARLVDIAPELSDTVAS
MAV_0515|M.avium_104 EAARVMADLIAEGAQTLTFVRSRRAAELTALGAQARLDEIAPELSAQVAS
MMAR_5141|M.marinum_M EAARVMADLIVEGAQTLTFVRSRRAAELTALGAQARLQDIAPQLSHTVAS
Mflv_1391|M.gilvum_PYR-GCK EAARVMADLIAEGARTLTFVRSRRGAELAALGARTRLADTAPDLADQVAS
Mvan_5399|M.vanbaalenii_PYR-1 EASRVMADLIAEGARTLTFVRSRRGAELAALGARARLADTAPELAQQVAS
MSMEG_6160|M.smegmatis_MC2_155 EAGRVMADLVAEGARTLTFVRSRRGAELTALGARARLSETAPDLVEQVAS
TH_0854|M.thermoresistible__bu EAARVMADLVAEGARTLTFVRSRRGAELTALGARSRLEEIAPELAQRVAS
MAB_0486c|M.abscessus_ATCC_199 ETSRLLADLVQEGARTLAFVRSRRGAEMTALGAAHRLSEVDPGLVSQVAA
*:.*::***: ***:**:******.**::**** ** : * * **:
Mb3673|M.bovis_AF2122/97 YRAGYLAEDRSALHQALAEGQLRGLATTNALELGVDIAGLDAVVLAGFPG
Rv3649|M.tuberculosis_H37Rv YRAGYLAEDRSALHQALAEGQLRGLATTNALELGVDIAGLDAVVLAGFPG
MAV_0515|M.avium_104 YRAGYLAEDRTALERALAEGRLRGLATTNALELGVDIAGLDAVVLAGFPG
MMAR_5141|M.marinum_M YRAGYLPEDRNCLEHALTEGRLRGLATTNALELGVDIAGLDAVVLAGFPG
Mflv_1391|M.gilvum_PYR-GCK YRAGYLAEDRRRLERALADGELRGMATTNALELGVDIAGLDAVVLAGFPG
Mvan_5399|M.vanbaalenii_PYR-1 YRAGYLAEDRRALERALAEGELRGVATTNALELGMDIAGLDAVVLAGFPG
MSMEG_6160|M.smegmatis_MC2_155 YRAGYLAEDRRELEHALSDGRLRGLATTNALELGIDIAGLDAVVMAGFPG
TH_0854|M.thermoresistible__bu YRAGYLAEDRRALEQALANGELRALATTNALELGIDIAGLDAVVLAGFPG
MAB_0486c|M.abscessus_ATCC_199 YRAGYLPEDRRALEQALSSGTLTGAATTNALELGVDIAGLDAVVVAGFPG
******.*** *.:**:.* * . *********:*********:*****
Mb3673|M.bovis_AF2122/97 TVASFWQQAGRSGRRGQGALVVLIARDDPLDTYLVHHPAALLDKPVERVV
Rv3649|M.tuberculosis_H37Rv TVASFWQQAGRSGRRGQGALVVLIARDDPLDTYLVHHPAALLDKPVERVV
MAV_0515|M.avium_104 TVASFWQQAGRSGRRGQGALVVLIARDDPLDTYLVHNPAALLGKPVERVV
MMAR_5141|M.marinum_M TVASFWQQAGRSGRRGQGALVVLIARDDPLDTYLVHHPAALLDKPVERVV
Mflv_1391|M.gilvum_PYR-GCK TVASFWQQAGRAGRRGQGALVVLIARDDPLDTYLVHHPAALLDKPIERVV
Mvan_5399|M.vanbaalenii_PYR-1 TVASFWQQAGRAGRRGQGALIVLIARDDPLDTYLVHHPAALLDKPIERVV
MSMEG_6160|M.smegmatis_MC2_155 TVASFWQQAGRSGRRGQGALVVLIARDDPLDTYLVHHPAALLDKPIERVV
TH_0854|M.thermoresistible__bu TVTSFWQQAGRSGRRGQSALIVLIARDDPLDTYLVHHPSALLDRPIERVV
MAB_0486c|M.abscessus_ATCC_199 TVASFWQQAGRSGRRGQGSLVVLVARDDPLDTYLVHHPEALLDKPIEAVV
**:********:*****.:*:**:************:* ***.:*:* **
Mb3673|M.bovis_AF2122/97 IDPVNPHLLGPQLLCAATELPLDDAEVRSWGAVEVAESLVDDGLLRRRNG
Rv3649|M.tuberculosis_H37Rv IDPVNPHLLGPQLLCAATELPLDDAEVRSWGAVEVAESLVDDGLLRRRNG
MAV_0515|M.avium_104 IDPRNPYILGPQLLCAATELPLQEAEVRELDATEVAEGLVDDGLLRRRNG
MMAR_5141|M.marinum_M IDPGNPYLLGPQLLCAATELPLDAVEVHELRAVEVVEALVDDGLLRRRGD
Mflv_1391|M.gilvum_PYR-GCK IDPANPYVLGPQLLCAATELPLTEAEVRMWDAESVAAGLVDDGLLRRRPS
Mvan_5399|M.vanbaalenii_PYR-1 IDPANPYVLGPQLLCAATELPLTDAEVRMWGAEPVAESLVDDGLLRRRPS
MSMEG_6160|M.smegmatis_MC2_155 IDPTNPHVLGPQLLCAAAELPLTEAEVRRWDAEAVAQRLVDDGLLRKRPA
TH_0854|M.thermoresistible__bu IDPTNPHVLGPQLLCAAAELPLTIEEVRRWDAEEVAESLVDDGLLRRRAS
MAB_0486c|M.abscessus_ATCC_199 IDPTNPHVLRPHLLCAATELPLTEDDVRSLDAGQVVSDLVDDGLLRKRPG
*** **::* *:*****:**** :*: * *. ********:*
Mb3673|M.bovis_AF2122/97 RYFPAPGVKPHAAVDVRGAIGGQIVIVEAGTGRLLGSVGVGQAPAAAHPG
Rv3649|M.tuberculosis_H37Rv RYFPAPGVKPHAAVDVRGAIGGQIVIVEAGTGRLLGSVGVGQAPAAAHPG
MAV_0515|M.avium_104 KYFPAPGLQPHAAVDIRGAGG-QVVILEADTGRLLGSAGAGQAPASVHPG
MMAR_5141|M.marinum_M KYFPAPGVQPHAAVDIRGSIGGQVVIVEADTGRLLGNVGADQAPASVHPG
Mflv_1391|M.gilvum_PYR-GCK GFFPAPGLDPHPAVDIRGSTGGQIAILEDGTGRMLGSTGAGQAPASVHPG
Mvan_5399|M.vanbaalenii_PYR-1 GFFPAPGLDPHPAVDIRGSTGGQIAILEAGTGRMLGSAGAGQAPASVHPG
MSMEG_6160|M.smegmatis_MC2_155 GYFPAPGVDPHPAVDIRGSTGGQIAILEADTGRMLGSAGAGQAASSVHPG
TH_0854|M.thermoresistible__bu GYFPSPGVDPHPAVDIRAAAGGQIAIVETGTGRLLGSAGVGQAPASVHPG
MAB_0486c|M.abscessus_ATCC_199 GYFAAPGLNPYAAVDIRGSIGGQVMILEADTGRVLGTVDTGRAPATVHPG
:*.:**:.*:.***:*.: * *: *:* .***:**.....:*.::.***
Mb3673|M.bovis_AF2122/97 AVYLHQGETYVVDSLDFQDGIAFVHAEDPGYATFAREVTDIAVTGTGERL
Rv3649|M.tuberculosis_H37Rv AVYLHQGETYVVDSLDFQDGIAFVHAEDPGYATFAREVTDIAVTGTGERL
MAV_0515|M.avium_104 AVYLHQGESYVVDSLDLQEGIAFVHAEDPGYATFAREITDIAVTGAGERL
MMAR_5141|M.marinum_M AVYLHQGESYVVDSLDTEEGLAFVHAADPGYATFARELTDIAVTGSGERL
Mflv_1391|M.gilvum_PYR-GCK AVYLHQGESYVVDSLDFEDGIAFVHAEDPGYTTFAREVTDISVTGAGERS
Mvan_5399|M.vanbaalenii_PYR-1 AVYLHQGESYVVDSLDFEDAVAFVHAEDPGYTTFARELTDINVTGAGERA
MSMEG_6160|M.smegmatis_MC2_155 AVYLHQGESYVVDSLDFEDGIAFVHAEDPGYTTFAREITDISVTGPGERV
TH_0854|M.thermoresistible__bu AVYLHQGESYLVDSLNFEEGVAFVHAEDPGYTTSAREITDISVTGVGERK
MAB_0486c|M.abscessus_ATCC_199 AVYLHSGETYVVDSLDFGDGIALVHNDRPDYSTFARSTIDIAITGEGERI
*****.**:*:****: :.:*:** *.*:* **. ** :** ***
Mb3673|M.bovis_AF2122/97 VFGPVALGLVPVTVTNHVVGYLRRQLSGEVLDFVELDMPEHTLPTTAVMY
Rv3649|M.tuberculosis_H37Rv VFGPVALGLVPVTVTNHVVGYLRRQLSGEVLDFVELDMPEHTLPTTAVMY
MAV_0515|M.avium_104 VFGPVTLGLVPVRVTHRVVGYLRRRLSGEVIDFIELDMPEHTLATTAAMY
MMAR_5141|M.marinum_M EFGPVTVGLVPVTVTHQVVGYLRRRLSGEVIDFVELDMPEHVLPTTAVMY
Mflv_1391|M.gilvum_PYR-GCK GHGPVTVGLVPVSVTNTVTGYLRRRLDGEVIDFIELDMPTRTLNTMAAMC
Mvan_5399|M.vanbaalenii_PYR-1 GYGPVTVGLVPVSVSNTVTGYLRRRMDGEVIDFVELDMPTRSLETMAVMC
MSMEG_6160|M.smegmatis_MC2_155 DHGPVTIGLVPVSVTNTVVGYLRRRMDGEVIDFVELDMPPRTLDTMAVMC
TH_0854|M.thermoresistible__bu TFGPVTIGLVPVSVTNTVTGYLRREMSGEVIDFVELDMPSRTLDTMAVMC
MAB_0486c|M.abscessus_ATCC_199 DLGDIQIGLVPVDVTHQVIGYLRTLTTGEILDFVELDMPSSTLSTKAVMY
* : :***** *:: * **** **::**:***** * * *.*
Mb3673|M.bovis_AF2122/97 TITSDALVRSGIEATRIPGSLHAAEHAAIGLLPLVASCDRGDIGGMSTAT
Rv3649|M.tuberculosis_H37Rv TITSDALVRSGIEATRIPGSLHAAEHAAIGLLPLVASCDRGDIGGMSTAT
MAV_0515|M.avium_104 TITEEALRDNGIDGPRIPGSLHAAEHAAIGLLPLVASCDRGDIGGLSTAA
MMAR_5141|M.marinum_M TITAEELLRNGVDAVAIPGSLHAAEHAAIGLLPLVASCDRGDIGGISTAI
Mflv_1391|M.gilvum_PYR-GCK TITPEALQDAGIDPLRIPGALHAAEHAAIGLLPLMASCDRGDIGGVSTAV
Mvan_5399|M.vanbaalenii_PYR-1 TITPEALQDNGIDPLRLPGSLHAAEHAAIGLLPLMASCDRGDIGGVSTAV
MSMEG_6160|M.smegmatis_MC2_155 TITPEALQDNGIEQLSVPGALHAAEHASIGLLPLVASCDRGDIGGVSTAV
TH_0854|M.thermoresistible__bu TITPEALADNDIDPVRIPGSLHAAEHAAIGLLPLVASCDRGDIGGVSTAV
MAB_0486c|M.abscessus_ATCC_199 TVTPEALAGIDIDPLSTPGALHAAEHAAIGLLPLVASCDRGDIGGVSTAL
*:* : * .:: **:*******:******:**********:***
Mb3673|M.bovis_AF2122/97 GPE--GLPSVFVYDGYPGGAGFAERGFRRARTWLGATAEAIEACECPSGC
Rv3649|M.tuberculosis_H37Rv GPE--GLPSVFVYDGYPGGAGFAERGFRRARTWLGATAEAIEACECPSGC
MAV_0515|M.avium_104 GPEPAGLPSVFVYDGHPGGAGFAERGFRRASTWLGATAAAIEACECPRGC
MMAR_5141|M.marinum_M GPE--GLPSVFVYDGYPGGAGFAERGFRRARTWLGATASAIEACECPSGC
Mflv_1391|M.gilvum_PYR-GCK GPV-DGLPTIFVYDGYPGGAGFAARGFRTLARWWEATASAIEACECPSGC
Mvan_5399|M.vanbaalenii_PYR-1 GPV-DGLPTIFVYDGYPGGAGFAARGFRTMDRWWEATASAIEACECPSGC
MSMEG_6160|M.smegmatis_MC2_155 GPV-DGLPSIFVYDGYPGGAGFADRGYRQLQTWWGATAAAIEACECPSGC
TH_0854|M.thermoresistible__bu GPD-SGLPTIFVYDGYPGGAGFADRGFDQFETWWGATADAIESCECPAGC
MAB_0486c|M.abscessus_ATCC_199 QPE-TGLPTVFVYDGHPGGAGFAARGYHKLATWLGATADAIDACECDAGC
* ***::*****:******* **: * *** **::*** **
Mb3673|M.bovis_AF2122/97 PSCVQSPKCGNGNDPLDKAGAVRVLRLVLAELSEESP
Rv3649|M.tuberculosis_H37Rv PSCVQSPKCGNGNDPLDKAGAVRVLRLVLAELSEESP
MAV_0515|M.avium_104 PSCVQSPKCGNGNDPLDKAGAARVLRLVLAELDGGPR
MMAR_5141|M.marinum_M PSCVQSPKCGNGNDPLDKAGAMQVLRLVLSQVGDDLL
Mflv_1391|M.gilvum_PYR-GCK PSCVQSPKCGNGNDPLDKDGAIRVLRLVIGALEHRHR
Mvan_5399|M.vanbaalenii_PYR-1 PSCVQSPKCGNGNDPLDKDGAILVLRMVVRLLTGRTD
MSMEG_6160|M.smegmatis_MC2_155 PSCVQSPKCGNGNDPLDKAGAVKVLRLVLGALTK---
TH_0854|M.thermoresistible__bu PSCVQSPKCGNGNDPLDKAGAVRVLRLVLDELEKGNT
MAB_0486c|M.abscessus_ATCC_199 PSCVQSPKCGNGNSPLDKRAAVKVLRLVLSRLPG---
*************.**** .* ***:*: :