For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SDRLTRRGGSPRSIESRGSMRPTRWRRSTHVAVGVAVLALVVAVVAAAALFTGKHSDAAEAVPPAPPPAT ADPGVVPVDLSAPTPTRRGLATALAAALANPDLGLITGRITDADTGAELWEQGARVPMQPASVNKVLTTA AALLTLDRDARLTTTVVAADDQPGLVVLRGGGDTTLSAAPKGTDTWYKGAARISDLADQVRARGIRVTRV RVDTSAYSGPTMAPGWDPADIDGGDIAPMESVMLDGGRTQPTTVESRRSKSPALDAGKALAAALGVEPES VTLMPSGMRGGTTIAEVQSAPLIERLRQMMNESDNVMAESIAREVAEALGRPQSFEGAVGAVLTQLRSVG IDTSGAKLVDSSGLSVDNRLTALTLDEVVNAAAGHTQPALRPLVDLLPIAGGSGTLSNRYLDTDAGRAAA GWLRAKTGSLTGTNALAGIVTDRSGRVLTFALISNNAGPTGRTAIDALAAVLRSCGCGA*
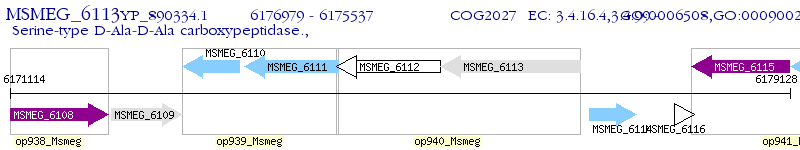
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6113 | dacB | - | 100% (480) | D-alanyl-D-alanine carboxypeptidase/D-alanyl-D-alanine-endopeptidase |
| M. smegmatis MC2 155 | MSMEG_4196 | - | 1e-05 | 29.33% (283) | LppL protein |
| M. smegmatis MC2 155 | MSMEG_1661 | - | 1e-05 | 26.71% (161) | D-alanyl-D-alanine carboxypeptidase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3651c | - | 1e-160 | 64.57% (460) | hypothetical protein Mb3651c |
| M. gilvum PYR-GCK | Mflv_1409 | - | 1e-178 | 70.43% (460) | D-alanyl-D-alanine |
| M. tuberculosis H37Rv | Rv3627c | - | 1e-160 | 64.78% (460) | hypothetical protein Rv3627c |
| M. leprae Br4923 | MLBr_00211 | - | 1e-145 | 60.39% (462) | putative carboxypeptidase |
| M. abscessus ATCC 19977 | MAB_0519 | - | 1e-156 | 63.20% (462) | peptidase S13 (D-alanyl-D-alanine carboxypeptidase |
| M. marinum M | MMAR_5127 | dacB1 | 1e-153 | 62.83% (460) | D-alanyl-D-alanine carboxypeptidase DacB1 |
| M. avium 104 | MAV_0529 | dacB | 1e-146 | 61.06% (452) | D-alanyl-D-alanine |
| M. thermoresistible (build 8) | TH_1412 | - | 1e-90 | 73.73% (236) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4203 | dacB1 | 1e-151 | 62.17% (460) | D-alanyl-D-alanine carboxypeptidase DacB1 |
| M. vanbaalenii PYR-1 | Mvan_5380 | - | 0.0 | 73.42% (459) | D-alanyl-D-alanine |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1409|M.gilvum_PYR-GCK --------------------MRPTGWRRSTYVVVG-AVVALLVVVLVAAA
Mvan_5380|M.vanbaalenii_PYR-1 --------------------MRPTRWRRSTYLVVGVSVLLLVVVVVVAAA
TH_1412|M.thermoresistible__bu --------------------------------------------------
MSMEG_6113|M.smegmatis_MC2_155 MSDRLTRRGGSPRSIESRGSMRPTRWRRSTHVAVGVAVLAL-VVAVVAAA
MMAR_5127|M.marinum_M --------------------MGPKRWRKSTQVFLGVAVLAFVAAVVAAAA
MUL_4203|M.ulcerans_Agy99 --------------------MGPKRWRKSTQVFLGVAVLAFVAAVVAAAA
Mb3651c|M.bovis_AF2122/97 --------------------MGPTRWRKSTHVVVGAAVLAFVAVVVAAAA
Rv3627c|M.tuberculosis_H37Rv --------------------MGPTRWRKSTHVVVGAAVLAFVAVVVAAAA
MLBr_00211|M.leprae_Br4923 --------------------MNPIRWQKS-YVVVGLAMLAFIIAAVKAAT
MAV_0529|M.avium_104 -------------------------------MFIGLGVLAFVAAVVVAAT
MAB_0519|M.abscessus_ATCC_1997 --------------MVTARALDPRHWRRSTHALLAVAVVLAIAALVLLAS
Mflv_1409|M.gilvum_PYR-GCK ALVAGRESDE-NVAVDPVPAPATADPGVIAVSDSADVPTERGLADALRGV
Mvan_5380|M.vanbaalenii_PYR-1 ALVATRQPAE-QPAVEAAPAPATAAPGVVPVSDSADKPTPQGLTRALQAV
TH_1412|M.thermoresistible__bu --------------------------------------------------
MSMEG_6113|M.smegmatis_MC2_155 ALFTGKHSDA-AEAVPPAPPPATADPGVVPVDLSAPTPTRRGLATALAAA
MMAR_5127|M.marinum_M FFTVGGRAAS-ARLVVPPPRPPTVKPAVTAVADTGVVPNSGAMAAALAQA
MUL_4203|M.ulcerans_Agy99 FFTVGGRAAS-ARLVVPPPRPPTVKPAVTAVADTGVVPNSGAMAAALAQA
Mb3651c|M.bovis_AF2122/97 LVTTGGHRAG-VRAPVPPPRPPTVKAGVVPVADTAATPSAAGVTAALAVV
Rv3627c|M.tuberculosis_H37Rv LVTTGGHRAG-VRAPAPPPRPPTVKAGVVPVADTAATPSAAGVTAALAVV
MLBr_00211|M.leprae_Br4923 VFTTGGHSTNGARSAVPPPRASTVKPALVPVTETAVVPSVGGLAAALASA
MAV_0529|M.avium_104 FFTTTGHGAGNAHAPIPAPHAPTVKPGMLPVSDTAELPTGPGVSAALAPV
MAB_0519|M.abscessus_ATCC_1997 -LTTGASTAHQLQSADPEPALITPAPGVVPVSDSAPIPTADGLAQALDRA
Mflv_1409|M.gilvum_PYR-GCK AADPNLGRFTGRITDALTGKQLWAQGADVPMQPASTNKLLTAAAALVTLD
Mvan_5380|M.vanbaalenii_PYR-1 LANPDLGRFTGRITDARSGDQLWAQGAGVPMQPASTNKMLTAAAALLTLD
TH_1412|M.thermoresistible__bu --------------------------------------------------
MSMEG_6113|M.smegmatis_MC2_155 LANPDLGLITGRITDADTGAELWEQGARVPMQPASVNKVLTTAAALLTLD
MMAR_5127|M.marinum_M AADPNLGKLGGRVSDALTGQVLWQQLDDVPLVPASTNKVLTATAALLTLD
MUL_4203|M.ulcerans_Agy99 AADPNLGKLGGRVSDALTGQVLWQQLDDVPLVPASTSKVLTATAALLTLD
Mb3651c|M.bovis_AF2122/97 AVDPDLGKLAGRITDALTGQELWQRLDDVPLVPASTNKILTAAAALLTLD
Rv3627c|M.tuberculosis_H37Rv AADPDLGKLAGRITDALTGQELWQRLDDVPLVPASTNKILTAAAALLTLD
MLBr_00211|M.leprae_Br4923 LADPSLGSFGGRITDAMTAKELWQQQDDMPLVPASANKLLTAAAALLTLD
MAV_0529|M.avium_104 AADPNLGRLGGRITDAITGKELWQVADDMPLVPASTNKVLTAAAALLTLD
MAB_0519|M.abscessus_ATCC_1997 LSDPALGMFTGRVTDALTGRELWQQGSTVPMVPASTNKVLTAAAALLTLE
Mflv_1409|M.gilvum_PYR-GCK RDARLTTRVVSTS---PGVIVLVAGGDQTLSAAPAKEDTWYRDAARISDL
Mvan_5380|M.vanbaalenii_PYR-1 RDARLTTRVLSAS---PGVVVLVGGGDPTLSAAPRNVDTWYRDAARIVDL
TH_1412|M.thermoresistible__bu --------------------------------------------------
MSMEG_6113|M.smegmatis_MC2_155 RDARLTTTVVAADDQ-PGLVVLRGGGDTTLSAAPKGTDTWYKGAARISDL
MMAR_5127|M.marinum_M RQARISTRVVAGSPSGQGPVVLVGAGDPTLSAAPPGVETWYRGAARISDL
MUL_4203|M.ulcerans_Agy99 RQARISTRVVAGSPSGQGSVVLVGAGDPTLSAAPPGVETWYRGAARISDL
Mb3651c|M.bovis_AF2122/97 RQARISTRVVAGGQNPQGPVVLVGAGDPTLSAAPPGQDTWYHGAARIGDL
Rv3627c|M.tuberculosis_H37Rv RQARISTRVVAGGQNPQGPVVLVGAGDPTLSAAPPGQDTWYHGAARIGDL
MLBr_00211|M.leprae_Br4923 RQTRISTRVVAAGPNAQGPVVLVGAGDPTLSAASPDQSTWYRGAPRISDL
MAV_0529|M.avium_104 RQARISTRVVAGSQNAQGPVVLVGAGDPVLSAAPPGQPTWYRGAARISDL
MAB_0519|M.abscessus_ATCC_1997 RDAKLTTSVVADTAGQRGLVTLVGGGDPIVSAAPAGTDTWYRDAARISDL
Mflv_1409|M.gilvum_PYR-GCK ADQVRRSGVDVRSVQVDVSFYTGPTMAPGWDPLDIDGGDIAPMEAIMLDG
Mvan_5380|M.vanbaalenii_PYR-1 ADQIRRSGVDVKSVQVDVSAYSGPSMAPGWDPLDIDGGDIAPMEAIMLDG
TH_1412|M.thermoresistible__bu ---------------------------------------------VMLDG
MSMEG_6113|M.smegmatis_MC2_155 ADQVRARGIRVTRVRVDTSAYSGPTMAPGWDPADIDGGDIAPMESVMLDG
MMAR_5127|M.marinum_M VDQIRRSGMTPTSVQVDISAFTGPTMAPGWDPADVGNGDIAPIEAAMIDA
MUL_4203|M.ulcerans_Agy99 VDQIRRSGMTPTSVQVDISAFTGPTMAPGWDPSDVGNGDIAPIEAAMIDA
Mb3651c|M.bovis_AF2122/97 VEQIRRSGVTPTAVQVDASAFSGPTMAPGWDPADIDNGDIAPIEAAMIDA
Rv3627c|M.tuberculosis_H37Rv VEQIRRSGVTPTAVQVDASAFSGPTMAPGWDPADIDNGDIAPIEAAMIDA
MLBr_00211|M.leprae_Br4923 VEQVRRSGVTPTAVQVDTSLFTGPTMAQGWDPADVDNGYTAPIESAMIDA
MAV_0529|M.avium_104 VEQIRRSGVTPTAVQVDTSAFSGPTMAQGWDPSDVDNGDIAPIESVMIDA
MAB_0519|M.abscessus_ATCC_1997 ADQVRKSGVAVTSITVDISRFGGPSMAPGWDPADIEGGDVAPIQAVMLDA
*:*.
Mflv_1409|M.gilvum_PYR-GCK GRTQPVSVESRRSRTPALDAGRALATALRLDPAKVTVLPPGRSAPDG-DE
Mvan_5380|M.vanbaalenii_PYR-1 GRTQPVTVESRRSRTPALDAGRALAVALRVDPAKVTVLP---SAARG-TE
TH_1412|M.thermoresistible__bu GRTQPVSEESRRSHTPALDAGRALAAALDVDPATVTVLP---SAYRGGTE
MSMEG_6113|M.smegmatis_MC2_155 GRTQPTTVESRRSKSPALDAGKALAAALGVEPESVTLMP---SGMRGGTT
MMAR_5127|M.marinum_M GRVQPTTVNSRRSLTPALDAGRELARALGLDPATVSIAS----APAGARQ
MUL_4203|M.ulcerans_Agy99 GRVQPTTVNSRRSLTPALDAGCELARALGLDPATVSIAS----APAGARQ
Mb3651c|M.bovis_AF2122/97 GRIQPTTVNSRRSRTPALDAGRELAKALGLDPAAVTIAS----APAGARQ
Rv3627c|M.tuberculosis_H37Rv GRIQPTTVNSRRSRTPALDAGRELAKALGLDPAAVTIAS----APAGARQ
MLBr_00211|M.leprae_Br4923 GRIQPTTVKSRRSRTPALDAGRELAKALGVAPDAVTIVK----ASSGARQ
MAV_0529|M.avium_104 GRIQPTTVNSRRTRTPALDAGRELAKALGLDPNAVTVAT----APSGARQ
MAB_0519|M.abscessus_ATCC_1997 GRTQPTDYDSRRSTTPALDAGRALAAALGVDPEAVRLDA----APPTAKS
** **. .***: :****** ** ** : * * : .
Mflv_1409|M.gilvum_PYR-GCK IASVQSAPLMERLRQMMHASDNVMAESIGREVADARNRPRSFAGAAQAVE
Mvan_5380|M.vanbaalenii_PYR-1 IASVQSPPLMQRLRDMMNFSDNVMAEAIGREVAAEVGRPQSFDGAARAVL
TH_1412|M.thermoresistible__bu IAAVESAPLIERLRVMMYASDNVMAETIGREVAAATGRPQSFAGAVEAVL
MSMEG_6113|M.smegmatis_MC2_155 IAEVQSAPLIERLRQMMNESDNVMAESIAREVAEALGRPQSFEGAVGAVL
MMAR_5127|M.marinum_M LAVVQSAPLIERLSQMMNASDNVMAECIAREVAAAINRPRSFSGAVDAVT
MUL_4203|M.ulcerans_Agy99 LAVVQSAPLIERLSQMMNASDNVMAECIAREVAAAINRPRSFSGAVDAVT
Mb3651c|M.bovis_AF2122/97 LAVVQSAPLIQRLSQMMNASDNVMAECIGREVAVAINRPQSFSGAVDAVT
Rv3627c|M.tuberculosis_H37Rv LAVVQSAPLIQRLSQMMNASDNVMAECIGREVAVAINRPQSFSGAVDAVT
MLBr_00211|M.leprae_Br4923 LAVVQSAPLVQRLSEMMDNSDNVLAECIGREVAAAINRPLSFAGAVDAVT
MAV_0529|M.avium_104 LAVVQSAPLVQRLSEMMDHSDNVLAECIAREVAAAINRPRSFAGAADAVT
MAB_0519|M.abscessus_ATCC_1997 IASVQSAPLMERLREMMNASDNVMAETIGREVALATGRAQTFSGTVDAVT
:* *:*.**::** ** ****:** *.**** .*. :* *:. **
Mflv_1409|M.gilvum_PYR-GCK DALDDAGIDTSGARLLDSSGLSVDNRLTAETLDEIIQAAAGESTPALRPL
Mvan_5380|M.vanbaalenii_PYR-1 STLEDAGIDTAGARLLDSSGLSVDNRLTAETLDEVVQAAAGDSEPALRPL
TH_1412|M.thermoresistible__bu DTLGDAGIDIDKAELFDNSGLSVDDRLTADTIDSVLAAAVGDDHAELRPL
MSMEG_6113|M.smegmatis_MC2_155 TQLRSVGIDTSGAKLVDSSGLSVDNRLTALTLDEVVNAAAGHTQPALRPL
MMAR_5127|M.marinum_M TRLNTAHINTTGAGLVDSSGLSVDDRLTARTLDDAIQAAAGPDQPALRPL
MUL_4203|M.ulcerans_Agy99 TRLNTALINTTGAGLVDSSGLSVDDRLTARTLDDAIQAAAGPDQPALRPL
Mb3651c|M.bovis_AF2122/97 SRLNTAHIDTAGAALVDSSGLSLDNRLTARTLDATMQAAAGPDQPALRPL
Rv3627c|M.tuberculosis_H37Rv SRLNTAHIDTAGAALVDSSGLSLDNRLTARTLDATMQAAAGPDQPALRPL
MLBr_00211|M.leprae_Br4923 NRLGTAHIDTTGAALVDSSGLSVNNRLTAKTLGGAVQAAAGPDQPVLRAL
MAV_0529|M.avium_104 NRLSTAHVDTAGATLMDSSGLSVDDRLTAKTLDGVLQAAAGPDQPALRAL
MAB_0519|M.abscessus_ATCC_1997 SQLRSAGIDLTDLTLRDSSGLSVDDRVTARTLDEVIGAAAGPDKPKLRPL
* . :: * *.****:::*:** *:. : **.* . **.*
Mflv_1409|M.gilvum_PYR-GCK VDLLAIAGGSGTMSNRYHDTEPGRAALGYLRAKTGSLTGTNTLAGIVTDE
Mvan_5380|M.vanbaalenii_PYR-1 VDLLPIAGGSGTLSNRYLDTDDGRAAAGYLRAKTGSLTGTNSLAGIVTDE
TH_1412|M.thermoresistible__bu VDMLPVAGGSGTLSYRYHDTAAGRAATGWLRAKTGSLTGTNALAGIVTGV
MSMEG_6113|M.smegmatis_MC2_155 VDLLPIAGGSGTLSNRYLDTDAGRAAAGWLRAKTGSLTGTNALAGIVTDR
MMAR_5127|M.marinum_M LDLLPIAGGSGTLGERFLDRATNLGPAGWLRAKTGSLTAINSLVGVVTDS
MUL_4203|M.ulcerans_Agy99 LDLLPIASGSGTLGERFLDRATNLGPAGWLRAKTGSLTAINSLVGVVTDS
Mb3651c|M.bovis_AF2122/97 LDLLPIAGGSGTLGERFLDAATDQGPAGWLRAKTGSLTAINSLVGVLTDR
Rv3627c|M.tuberculosis_H37Rv LDLLPIAGGSGTLGERFLDAATDQGPAGWLRAKTGSLTAINSLVGVLTDR
MLBr_00211|M.leprae_Br4923 LDLLPIAGGSGTLSDRFLSSATHQGPAGWLRAKTGSLTAINSLVGVVTDR
MAV_0529|M.avium_104 LDLLPIAAGSGTLGDRFTDASTNQGPAGWLRAKTGSLTAINALAGVLTDR
MAB_0519|M.abscessus_ATCC_1997 LDVLPIAGGSGTLSERFVTQN--QTSAGWLRAKTGSLTGVNTLAGVVTDV
:*:*.:*.****:. *: . *:*********. *:*.*::*.
Mflv_1409|M.gilvum_PYR-GCK SGRVLTFALLSNDA-GPTGRTSLDAFAATLRSCGCR-
Mvan_5380|M.vanbaalenii_PYR-1 SGRVLTFALLSNDA-GPTGRTALDAFAATLRSCGCR-
TH_1412|M.thermoresistible__bu SGRVLTFTLISNDA-GPMGRTALDAVAAVLRSCGCRT
MSMEG_6113|M.smegmatis_MC2_155 SGRVLTFALISNNA-GPTGRTAIDALAAVLRSCGCGA
MMAR_5127|M.marinum_M SGRVLTFAFISNDA-GPNGRNAMDALATTLWSCGCTT
MUL_4203|M.ulcerans_Agy99 SGRVLTFAFISNDA-GPNGRNAMDALATTLWSCGCTT
Mb3651c|M.bovis_AF2122/97 SGRVLTFAFISNEA-GPNGRNAMDALATKLWFCGCTT
Rv3627c|M.tuberculosis_H37Rv SGRVLTFAFISNEA-GPNGRNAMDALATKLWFCGCTT
MLBr_00211|M.leprae_Br4923 SGRVLTFAFISNDA-GPTGRAAMDELATRLWMCGCAT
MAV_0529|M.avium_104 SGRVLTFAFISNDA-GPNGRNAMDALATRLWFCGCS-
MAB_0519|M.abscessus_ATCC_1997 SGRVLTFSLMQNHATAPTARNAVDNTAAVLRSCGCS-
*******:::.*.* .* .* ::* *: * ***