For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TVRSRLPYALVVGALVVLLLLVMTAGVAIGSVPIAPRQVWQIVLNGIHPALAEQTWPAVRTSIVLDARLP RVVLAAVVGAGLAACGMVLQAVVRNPLADPMLLGVSSGATVGAVAVLVAGAGVRQLVTLPVAAFVGALAA LVAVYFLARSGGRMTTVRLILAGVAVAEVLSAVASLLIVTSDDPHKAQAALRWMLGGLGGATWRAVWIPA VVVLIGVVVLLAVTRPLNLLYTGEEAAASLGLDVHRFRAAMFVVVALMVGAMVAVSGSIGFVGLIMPHAV RFLVGADHRRALPAVALLGAAFLVVCDIAARTVGAPEELPVGVLTALVGGPFFLWLMRRRAPA*
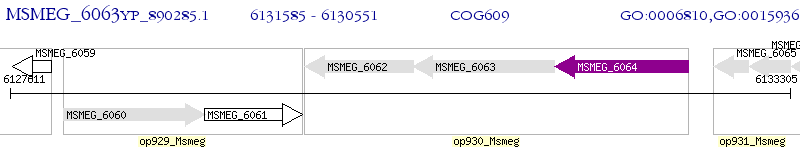
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6063 | - | - | 100% (344) | Fe uptake system integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_4559 | - | 5e-72 | 47.01% (334) | ABC transporter, membrane spanning protein |
| M. smegmatis MC2 155 | MSMEG_0012 | - | 2e-49 | 35.84% (332) | ferric enterobactin transport system permease protein FepD |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3883 | - | 1e-108 | 59.41% (340) | transport system permease protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00335 | - | 6e-07 | 24.47% (282) | putative ABC-transporter transmembrane protein |
| M. abscessus ATCC 19977 | MAB_0549c | - | 8e-75 | 45.54% (336) | putative heme/hemin ABC transporter, permease protein |
| M. marinum M | MMAR_1101 | - | 9e-05 | 26.75% (228) | hypothetical protein MMAR_1101 |
| M. avium 104 | MAV_0581 | - | 1e-08 | 26.75% (243) | ABC-type Mn2+/Zn2+ transport system, permease component |
| M. thermoresistible (build 8) | TH_3095 | - | 2e-67 | 44.64% (336) | PUTATIVE putative ABC transporter, membrane spanning protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1828 | - | 5e-05 | 26.13% (199) | hypothetical protein Mvan_1828 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6063|M.smegmatis_MC2_155 -------------------------------------------MT--VRS
Mflv_3883|M.gilvum_PYR-GCK -------------------------------------------MTRPVRA
MAB_0549c|M.abscessus_ATCC_199 -------------------------------------------MPTAQHT
TH_3095|M.thermoresistible__bu ----------------------------------------------VSLA
MLBr_00335|M.leprae_Br4923 --------------------------------------------------
MAV_0581|M.avium_104 --------------------------------------------------
MMAR_1101|M.marinum_M MLSGSEPGLCRVAVHSGTAVVDLSLPAALPVAELIPAIVESLGGRDIGAA
Mvan_1828|M.vanbaalenii_PYR-1 -----------------------------------------------MIA
MSMEG_6063|M.smegmatis_MC2_155 RLPYALVVGALVVLLLLVMTAGVAIGS-VPIAPRQVWQIVLNGIHPALAE
Mflv_3883|M.gilvum_PYR-GCK SLPYPVLMLGLAATVLAAVVAGLAIGS-IAIPPGDVARILAHQIVPNLVD
MAB_0549c|M.abscessus_ATCC_199 RSLILAVLGGLAALVLLTVTA-LAFGT-ADVPWRDCLHFLWAGASGGSVP
TH_3095|M.thermoresistible__bu HRAWLLGAFGLAALLASIAVA-VTIGP-STVGAGEVWAVVRARLAGP--P
MLBr_00335|M.leprae_Br4923 ----------------------------MDDRLAHMWNHLFS------FD
MAV_0581|M.avium_104 ----------------------------MEHRLADMADHLFS------FD
MMAR_1101|M.marinum_M PTRYQLCPLGAGALPPSQSLAQSGIRDGTVLSLSRCSAEPAPGPHDDDAE
Mvan_1828|M.vanbaalenii_PYR-1 LTIGLAVFVGIALGLLGGGGSILTVPLLAYVAGMDAKAAIATSLLVVGVT
MSMEG_6063|M.smegmatis_MC2_155 QTWPAVRTSIVLDARLPRVVLAAVVGAGLAACGMVLQAVVRNPLADPMLL
Mflv_3883|M.gilvum_PYR-GCK PTWPAHFQTVITQVRGPRVVLGAVVGAGLAVVGMTLQALVRNPLADPYLL
MAB_0549c|M.abscessus_ATCC_199 AVDAANYR-IVVESRLPRVLLAVLVGAGLSVVGVIVQAMVRNALADPYVL
TH_3095|M.thermoresistible__bu SGLTRIQEGIVWDLRLPRSLLAAACGAGLAVCGVILQSMLRNPLADPFLL
MLBr_00335|M.leprae_Br4923 ITTHLLSHNFVQQALLAALLLGLVAG--------LIGPFIVMRQMSFAVH
MAV_0581|M.avium_104 ITVHLLGHDFVQQALVAAALLGLVAG--------LIGPFIVMRQMSFAVH
MMAR_1101|M.marinum_M AVLATLRGSTRPPSRRAKELTGALTAGGVGGVGAVLLVRDALGNTDRHGA
Mvan_1828|M.vanbaalenii_PYR-1 SAIGAISHARAGRVQWRTGLIFGAAG---------MAGAYAGGLLARYIP
: . :
MSMEG_6063|M.smegmatis_MC2_155 GVSSGATVG-AVAVLVAGAG-VRQLVTLPVAAF---------VGALAALV
Mflv_3883|M.gilvum_PYR-GCK GVSSGASVG-AVAVIVTGVS-LVGALSTSAAAF---------AGALLACS
MAB_0549c|M.abscessus_ATCC_199 GISSGASVG-ATAVVVYGVLGTLGTYALSLSAS---------LGALAATA
TH_3095|M.thermoresistible__bu GVSSGASTG-AVSIVVLGVG--AGSIGLAGGAF---------IGAVGAFG
MLBr_00335|M.leprae_Br4923 GSSELSLTG-ATFALLAGFE-------MGVGAL---------VGSALAAT
MAV_0581|M.avium_104 GSSELSLTG-AAFALLVGIG-------VGVGAL---------IGSALAAA
MMAR_1101|M.marinum_M ATAVVAITSGVLALLVAGMARRTGREPTAVFTLNVIATTFVAVGALLAVP
Mvan_1828|M.vanbaalenii_PYR-1 G--TILLIGFAVMMIATAIAMLRGRKTIETGET----------GHRLPVP
. . . : . * .
MSMEG_6063|M.smegmatis_MC2_155 --------AVYFLARSGGRMTTVRLI---------------------LAG
Mflv_3883|M.gilvum_PYR-GCK --------LVYGLSRAGGRITTTRLV---------------------LAG
MAB_0549c|M.abscessus_ATCC_199 --------VVYQIARTPQGLVPLRLV---------------------LVG
TH_3095|M.thermoresistible__bu --------LVLLLTVAAGGG-TDRVI---------------------LAG
MLBr_00335|M.leprae_Br4923 --------LFGILGRR----ARERDS---------------------VIG
MAV_0581|M.avium_104 --------LFGVLGRR----ARERDS---------------------VIG
MMAR_1101|M.marinum_M GRLGVPNLMLAAMGASVAAALAIRLTGCGGVTFTAVMCCAIVIAAAALCG
Mvan_1828|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6063|M.smegmatis_MC2_155 VAVAEVLSAVASLLIVTSDDPHKAQAALRWMLG----GLGGATWRAVWIP
Mflv_3883|M.gilvum_PYR-GCK VAVAYVLSAVTSLMLITADGGDRARQVLTWLLG----GLGGARWDNLWLP
MAB_0549c|M.abscessus_ATCC_199 TAVGYGLSAVTTVLVFLAPNGEAARSVMFWLLG----SLGASSWDKVPVA
TH_3095|M.thermoresistible__bu VATTQLFSALTSFIVFTAADAEQTRGVLFWLLG----SLASASWRDVAVV
MLBr_00335|M.leprae_Br4923 VVLAFGLGLAVLFIHLYPG---RTGTSLALLTG----QIIDVGYSGLATL
MAV_0581|M.avium_104 VVLAFGLGLAVLFIHLYPG---RTATSFALLTG----QIVGVGYTGLTML
MMAR_1101|M.marinum_M VITGAALYVVGSLTTLAALVALEVSARVSMVVAGLSPKLPAAPGSEEAPE
Mvan_1828|M.vanbaalenii_PYR-1 KIVAEGLIVGLVTGLVGAGGGFLVVPALALLGG-------------LPMP
: . . . . : .
MSMEG_6063|M.smegmatis_MC2_155 AVVVLIGVVVLLAVTRPLNLLYTGEEAAASLGLDVHRFRAAMFVVVALMV
Mflv_3883|M.gilvum_PYR-GCK LAVVGAGLVVLMAHGRTLNVLLAGDEAAATLGVDTQRFRVRAFVLTSLVT
MAB_0549c|M.abscessus_ATCC_199 LAATVIGILCAGGLARQLNALAMGDEVSASLGLDAGRFRVALFILSAAMT
TH_3095|M.thermoresistible__bu TALVAAGLVACLVWASSLDSFTFGEDAAAGLGVRVGPTRLGLLLVTASMT
MLBr_00335|M.leprae_Br4923 ALVCLFVIAVLTTCYRPLLFATVDPEVAAACGVPVHALGIVFAALVGLVA
MAV_0581|M.avium_104 ALVCLLVIAVLATCYRPLLFATVDPDVAAARGVPVHALGIVFAALVGVVA
MMAR_1101|M.marinum_M LAAPDLLATRAVRADRWLNGLHGGFSSAAALGAVVAVLGSQRAIGLTAVT
Mvan_1828|M.vanbaalenii_PYR-1 IAVGTSLIVIAMKSFAGLAGYLTSVHINWTLALAVTAAAVIGALIGARLT
* . . . :.
MSMEG_6063|M.smegmatis_MC2_155 GAMVAVSG----------SIGFVGLIMPHAVRFLVGADHRRALPAVALLG
Mflv_3883|M.gilvum_PYR-GCK GVLVAVSG----------PIGFVGLILPHAVRLLVGSDHRRALPAAALAG
MAB_0549c|M.abscessus_ATCC_199 GCFVSICG----------AVGFVGLVIPHAARLLVGADHRRVLIVAPVLG
TH_3095|M.thermoresistible__bu AVIVSTSG----------AIGFVGLVLPHVARALVGVAHRRLLPLSAVIG
MLBr_00335|M.leprae_Br4923 AQAVQVVG----------ALLVMSLLITPAAAAAQVVASPVAAMVAAVIF
MAV_0581|M.avium_104 AQAVQIVG----------ALLVMSLLITPAAAAARVVASPGAAMLASVAF
MMAR_1101|M.marinum_M GAVLLLRARAQVDGRSRLMFLIAGYVAGTAAFAVAAPGLAQHGPWIAILT
Mvan_1828|M.vanbaalenii_PYR-1 AMINPDSLR----------KTFGWFVLVMSSVILAQEIH----PAVGITA
. . : :
MSMEG_6063|M.smegmatis_MC2_155 AAFLVVCDIAARTVGAPEELPVGV--------LTALVGGPFFLWLMRRRA
Mflv_3883|M.gilvum_PYR-GCK AGFLVLADIAARTWASPQEIPVGV--------LTALCGGPFFLWLLRRDA
MAB_0549c|M.abscessus_ATCC_199 ACFLVAADLVARTAIPPQELPLGA--------ITAAVGVPVFVGLMRRRA
TH_3095|M.thermoresistible__bu AVFLVWVDTVARVIIDPQELPVGV--------ATALIGVPVFALILLRRR
MLBr_00335|M.leprae_Br4923 AEVSAVGGVVLSLAPG-VPVSVFV--------ATISFLIYLFCWLLGRHR
MAV_0581|M.avium_104 AEVSALGGIVLSLAPG-VPVSVFV--------ATISFLIYLACWLIGRRR
MMAR_1101|M.marinum_M AALTALALHLGFIAPTRPPSPIACRLIDALEFSASASLLPLACWTCGVFS
Mvan_1828|M.vanbaalenii_PYR-1 AALTLIAAATTLACQRYAHCPLSR----------------LTTRLTTPKA
* . .: .
MSMEG_6063|M.smegmatis_MC2_155 PA--------
Mflv_3883|M.gilvum_PYR-GCK RRADTGAAW-
MAB_0549c|M.abscessus_ATCC_199 MSAGAVG---
TH_3095|M.thermoresistible__bu RAV-------
MLBr_00335|M.leprae_Br4923 EVCA------
MAV_0581|M.avium_104 EAAT------
MMAR_1101|M.marinum_M AVGGLGLTWT
Mvan_1828|M.vanbaalenii_PYR-1 AA--------