For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTITSTTVEPGIVSVTVNYPPVNAVPSRGWFELADAITAAGRDPQTHVVILRAEGRGFNAGVDIKEMQNT EGFTALIDANRGCFAAFKAVYECEVPVVTAVNGFCVGGGIGLVGNSDVIVASDDAKFGLPEVERGALGAA THLSRLVPQHMMRRLFFTAATVDAATLHHFGSVHEVVPREDLDEAALRVARDIATKDTRVIRAAKEALNF IDVQRVNASYRMEQGFTFELNLSGVSDEHRDAFAGTEKRAK
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
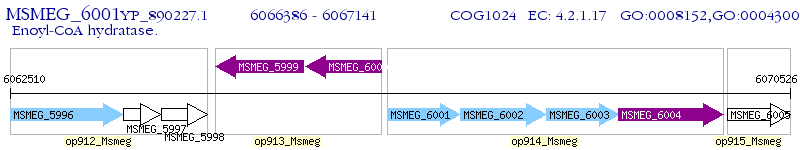
| M. smegmatis MC2 155 | MSMEG_6001 | - | - | 100% (251) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_5277 | - | 5e-23 | 32.67% (202) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_2880 | - | 2e-22 | 36.59% (205) | enoyl-CoA hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3580 | echA20 | 1e-121 | 87.65% (243) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_1490 | - | 1e-128 | 89.24% (251) | enoyl-CoA hydratase |
| M. tuberculosis H37Rv | Rv3550 | echA20 | 1e-121 | 87.65% (243) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 5e-23 | 34.17% (199) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_0606c | - | 1e-115 | 79.84% (248) | enoyl-CoA hydratase |
| M. marinum M | MMAR_5039 | echA20 | 1e-124 | 88.48% (243) | enoyl-CoA hydratase EchA20 |
| M. avium 104 | MAV_0610 | - | 1e-128 | 88.45% (251) | enoyl-CoA hydratase |
| M. thermoresistible (build 8) | TH_1755 | echA20 | 1e-130 | 91.24% (251) | PROBABLE ENOYL-CoA HYDRATASE ECHA20 (ENOYL HYDRASE) |
| M. ulcerans Agy99 | MUL_4113 | echA20 | 1e-124 | 88.07% (243) | enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_5266 | - | 1e-129 | 91.53% (248) | enoyl-CoA hydratase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3580|M.bovis_AF2122/97 MPITSTTPE--PGIVAVTVDYP-PVNAIPSKAWFDLADAVTAAGANSDTR
Rv3550|M.tuberculosis_H37Rv MPITSTTPE--PGIVAVTVDYP-PVNAIPSKAWFDLADAVTAAGANSDTR
MMAR_5039|M.marinum_M MTITSTTAE--PGIVAITVDYP-PVNAIPSRGWFELADALTAAGADLETR
MUL_4113|M.ulcerans_Agy99 MTITSTTAE--PGIVAITVDYP-PVNAIPSRGWFELADALTAAGADLETR
Mflv_1490|M.gilvum_PYR-GCK MPITTKTVE--PGIVAVTVDYP-PVNAIPSRGWFELADIITAAGRDQSTH
Mvan_5266|M.vanbaalenii_PYR-1 MPITTKTVE--PGIVSVTVDYP-PVNAIPSRGWFELADTITAAGRDHSTH
TH_1755|M.thermoresistible__bu MPITTTSPE--PGIVSVTVDYP-PVNAIPSRGWFELADAITAAGRDQQTH
MSMEG_6001|M.smegmatis_MC2_155 MTITSTTVE--PGIVSVTVNYP-PVNAVPSRGWFELADAITAAGRDPQTH
MAV_0610|M.avium_104 MPITSKTIE--PGIVAVTVDFP-PVNAIPSRGWFDLADTVTAAGQDPETH
MAB_0606c|M.abscessus_ATCC_199 MGITTATVE--PGIVTVTVDYP-KVNAIPSRGWFELADAILAAGNDPSTH
MLBr_02402|M.leprae_Br4923 MKYDTILVDGYQRVGIITLNRPQALNALNSQMMNEITNAAKELDIDPDVG
* : : : :*:: * :**: *: :::: . : ..
Mb3580|M.bovis_AF2122/97 AVILRAEGRGFNAGVDIKEMQRTEGFTALIDANRGCFAAFRAVYECAVPV
Rv3550|M.tuberculosis_H37Rv AVILRAEGRGFNAGVDIKEMQRTEGFTALIDANRGCFAAFRAVYECAVPV
MMAR_5039|M.marinum_M AVILRAEGRGFNAGVDIKEMQRTEGFTALIDANRGCFAAFRAVYECAVPV
MUL_4113|M.ulcerans_Agy99 AVILRADGRGFNAGVDIKEMQRTEGFTALIDANRGCFAAFRAVYECAVPV
Mflv_1490|M.gilvum_PYR-GCK VVILRAEGRGFNAGVDIKEMQNTEGFTALIDANRGCYEAFRAVYECAVPV
Mvan_5266|M.vanbaalenii_PYR-1 VVILRAEGRGFNAGVDIKEMQNTEGFTALIDANRGCYEAFRAVYECAVPV
TH_1755|M.thermoresistible__bu VVILRAEGRGFNAGVDIKEMQNTEGFTALIDANRGCYEAFRAVYECAVPV
MSMEG_6001|M.smegmatis_MC2_155 VVILRAEGRGFNAGVDIKEMQNTEGFTALIDANRGCFAAFKAVYECEVPV
MAV_0610|M.avium_104 VVILRAEGRGFNAGVDIKEMQRTEGFTALIDANRGCFAAFRAVYECAVPV
MAB_0606c|M.abscessus_ATCC_199 VVILRAEGRGFNAGADIKEMQATDGFTALIDTNRGCFEAFSAVYRCKVPV
MLBr_02402|M.leprae_Br4923 AILITGSPKVFAAGADIKEMASLT-FTDAFDAD--FFSAWGKLAAVRTPM
.::: .. : * **.***** ** :*:: : *: : .*:
Mb3580|M.bovis_AF2122/97 IAAVNGFCVGGGIGLVGNSDVIVASEDATFGLPEVERGAL---GAATHLS
Rv3550|M.tuberculosis_H37Rv IAAVNGFCVGGGIGLVGNSDVIVASEDATFGLPEVERGAL---GAATHLS
MMAR_5039|M.marinum_M IAAVNGFCVGGGIGLVGNSDVIVASEDATFGLPEVERGAL---GAATHLS
MUL_4113|M.ulcerans_Agy99 IAAVNGFCVGGGIGLVGNSDVIVASEDATFGLPEVERGAL---GAATHLS
Mflv_1490|M.gilvum_PYR-GCK VAAVNGFCVGGGIGLVGNADVIVASDDAKFGLPEVERGAL---GAATHLS
Mvan_5266|M.vanbaalenii_PYR-1 VAAVNGFCVGGGIGLVGNSDVIVASDDAKFGLPEVERGAL---GAATHLS
TH_1755|M.thermoresistible__bu VAAVNGFCVGGGIGLVGNSDVIVASDDAKFGLPEVERGAL---GAATHLS
MSMEG_6001|M.smegmatis_MC2_155 VTAVNGFCVGGGIGLVGNSDVIVASDDAKFGLPEVERGAL---GAATHLS
MAV_0610|M.avium_104 VAAVNGFCVGGGIGLVGNSDVIVASDDATFGLPEVERGAL---GAATHLS
MAB_0606c|M.abscessus_ATCC_199 IAAVNGFCLGGGIGLVGNADVIVAADDAVFSVPEVDRGAL---GTATHLQ
MLBr_02402|M.leprae_Br4923 IAAVAGYALGGGCELAMMCDLLIAADTAKFGQPEIKLGVLPGMGGSQRLT
::** *:.:*** *. .*:::*:: * *. **:. *.* * : :*
Mb3580|M.bovis_AF2122/97 RLVPQHLMRRLFFTAATVDAATLQHFGSVHEVVSRDQLDEAALRVARDIA
Rv3550|M.tuberculosis_H37Rv RLVPQHLMRRLFFTAATVDAATLQHFGSVHEVVSRDQLDEAALRVARDIA
MMAR_5039|M.marinum_M RLVPQHLMRRLFFTAATVDAATLHHFGSVHEVVPRAELDESALRVARDIA
MUL_4113|M.ulcerans_Agy99 RLVPQHLMRRLFFTAATVDAATLHHFGSVHEVVPRAELDESALRVARDIA
Mflv_1490|M.gilvum_PYR-GCK RLVPQHLMRRLFFTAATVGADTLHHFGSVHEVVPRADLDEAALRVARDIA
Mvan_5266|M.vanbaalenii_PYR-1 RLVPQHMMRRLFFTAATVTADTLHHFGSVHEVVPRADLDEAALRVARDIA
TH_1755|M.thermoresistible__bu RLVPQHMMRRLFFTAATVDAHTLHHFGSVHEVVPRAELDEAALRVARDIA
MSMEG_6001|M.smegmatis_MC2_155 RLVPQHMMRRLFFTAATVDAATLHHFGSVHEVVPREDLDEAALRVARDIA
MAV_0610|M.avium_104 RLVPQHLMRRLFFTAATVDAATLHHFGSVHEVVPRGELDDAALRVAREIA
MAB_0606c|M.abscessus_ATCC_199 RLVPQHVMRRMFYTAAKIDAKTLHHYGSIHEVVPRAELDEAALRVARDIA
MLBr_02402|M.leprae_Br4923 RAIGKAKAMDLILTGRTIDAAEAERSGLVSRVVLADDLLPEAKAVATTIS
* : : :: *. .: * .: * : .** :* * ** *:
Mb3580|M.bovis_AF2122/97 AKDTRVIRAAKEALNFIDVQRVNASYRMEQGFTFELNLAGVADEHRDAFV
Rv3550|M.tuberculosis_H37Rv AKDTRVIRAAKEALNFIDVQRVNASYRMEQGFTFELNLAGVADEHRDAFV
MMAR_5039|M.marinum_M AKDTRVIRAAKEALNLIDVQRVNSSYRMEQGFTFELNLAGVADEHRDAFV
MUL_4113|M.ulcerans_Agy99 AKDTRVIRAAKEALNLIDVQRVNSSYRMEQGFTFELNLAGVADEHRDAFV
Mflv_1490|M.gilvum_PYR-GCK SKDTRVIRAAKEALNFIDIQPVNARYRMEQGFTFELNLAGVSDEHRDAFA
Mvan_5266|M.vanbaalenii_PYR-1 SKDTRVIRAAKEALNFIDIQPVNARYRMEQGFTFELNLAGVSDEHRDAFA
TH_1755|M.thermoresistible__bu AKDTRVIRAAKEALNFIDVQRVNSSYRMEQGFTFELNLAGVADEHRDAFA
MSMEG_6001|M.smegmatis_MC2_155 TKDTRVIRAAKEALNFIDVQRVNASYRMEQGFTFELNLSGVSDEHRDAFA
MAV_0610|M.avium_104 AKDTRVIRAAKEALNFIDVQRVNSSYRMEQGFTFELNLAGVSDEHRDAFA
MAB_0606c|M.abscessus_ATCC_199 AKDTRVIRAAKEAINFIDPQDVESSYRMEQGFTFELNLAGVADEHRDAFV
MLBr_02402|M.leprae_Br4923 QMSRSATRMAKEAVNRSFESTLAEGLLHERRLFHSTFVTDDQSEGMAAFI
. . * ****:* . : *: : .. ::. .* **
Mb3580|M.bovis_AF2122/97 KKS-------
Rv3550|M.tuberculosis_H37Rv KKS-------
MMAR_5039|M.marinum_M KKS-------
MUL_4113|M.ulcerans_Agy99 KKS-------
Mflv_1490|M.gilvum_PYR-GCK GTDKGSK---
Mvan_5266|M.vanbaalenii_PYR-1 GTEKGKA---
TH_1755|M.thermoresistible__bu GTNKGKKQ--
MSMEG_6001|M.smegmatis_MC2_155 GTEKRAK---
MAV_0610|M.avium_104 GTQKGKK---
MAB_0606c|M.abscessus_ATCC_199 ETGKPKGN--
MLBr_02402|M.leprae_Br4923 EKRAPQFTHR
.