For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NRASKSGSSRSTYTSESGVVLLSVRSSHDADVRRASGCNITVHGIGATDRELDPGEHLTWVSVAQFEQVL DAVSGRSDVRITFDDGNASDVKVALPRLVERGITAEFFLLAGELGKLGRVDDTGVRELLDAGMSIGSHGW AHRDWRRIDDAQAHEEFEDAHERLSAVTGSSVSRVAIPFGSYDRHVLRRLRDAGVTRVYTSDGGRARPEA WLQARTSLRHDIDADWIYRLLDPHTPLVLSARNSAARAVKRWRG*
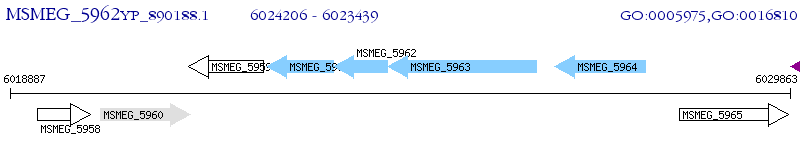
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5962 | - | - | 100% (255) | glycosyl transferase, family protein 2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1126 | - | 3e-08 | 32.97% (91) | glycosyl hydrolase |
| M. gilvum PYR-GCK | Mflv_2063 | - | 1e-10 | 29.53% (149) | polysaccharide deacetylase |
| M. tuberculosis H37Rv | Rv1096 | - | 3e-08 | 32.97% (91) | glycosyl hydrolase |
| M. leprae Br4923 | MLBr_01950 | - | 8e-08 | 27.47% (91) | carbohydrate degrading enzyme |
| M. abscessus ATCC 19977 | MAB_0459c | - | 2e-07 | 30.77% (91) | glycosylhydrolase |
| M. marinum M | MMAR_4371 | - | 1e-07 | 30.77% (91) | glycosyl hydrolase |
| M. avium 104 | MAV_1218 | - | 1e-07 | 28.83% (111) | carbohydrate degrading enzyme |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0184 | - | 9e-08 | 30.77% (91) | glycosyl hydrolase |
| M. vanbaalenii PYR-1 | Mvan_4651 | - | 4e-10 | 31.67% (120) | polysaccharide deacetylase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4371|M.marinum_M --MRKRPDNQAWRYWR--------------MVVGVVAAVAVLV--IGSLT
MUL_0184|M.ulcerans_Agy99 --MRKRPDDQAWRYWR--------------MVVGVVAAVAVLV--IGSLT
MAV_1218|M.avium_104 --MAKRPDTLAWRYWR--------------TVVGVLAAVAVLV--IGGLT
Mb1126|M.bovis_AF2122/97 --MPKRPDNQTWRYWR--------------TVTGVVVAGAVLV--VGGLS
Rv1096|M.tuberculosis_H37Rv --MPKRPDNQTWRYWR--------------TVTGVVVAGAVLV--VGGLS
MLBr_01950|M.leprae_Br4923 --MLKRPDTQAWRYSR--------------TVFGVVAAGAVLV--IGALT
MAB_0459c|M.abscessus_ATCC_199 --MGTVLDSVRFQIAA--------------VAVVLVAAAGVVG--IGVRH
Mflv_2063|M.gilvum_PYR-GCK --MTSESGALDDAHRR--------------QARPIRRLGAVVLGVITVFT
Mvan_4651|M.vanbaalenii_PYR-1 --MTSGSGAQYDTRGA--------------RARRIR-SAAILLGLVGGFI
MSMEG_5962|M.smegmatis_MC2_155 MNRASKSGSSRSTYTSESGVVLLSVRSSHDADVRRASGCNITVHGIGATD
. . : :
MMAR_4371|M.marinum_M GHV---TRADDQSCAVVK----------CVALTFDDGPSPFTDRLLQILK
MUL_0184|M.ulcerans_Agy99 GHV---TRADDQSCAVVK----------CVALTFDDGPSPFTDRLLQILK
MAV_1218|M.avium_104 GHV---TRADDLSCSVVK----------CVALTFDDGPGPFDERLLQILK
Mb1126|M.bovis_AF2122/97 GRV---TRAENLSCSVIK----------CVALTFDDGPGPYTDRLLHILT
Rv1096|M.tuberculosis_H37Rv GRV---TRAENLSCSVIK----------CVALTFDDGPGPYTDRLLHILT
MLBr_01950|M.leprae_Br4923 GHV---TRADDMSCAQVK----------CIALTFDDGPGPYTDRLVQILK
MAB_0459c|M.abscessus_ATCC_199 RLL---LGPDTVDCSRYK----------CVALTFDDGPTPFTDRLLQTLT
Mflv_2063|M.gilvum_PYR-GCK AAP--TAAAGPVDCAAVP----------CVALTFDDGPGPHTDRLLDVLR
Mvan_4651|M.vanbaalenii_PYR-1 AAVPAPAAADSVDCARVK----------CVALTFDDGPGPFTDRLLQTLR
MSMEG_5962|M.smegmatis_MC2_155 RELDPGEHLTWVSVAQFEQVLDAVSGRSDVRITFDDGNASDVKVALPRLV
. : : :***** . . : *
MMAR_4371|M.marinum_M DNDAKATFFLIGNKV----AANPAAAKRIAEAGMEIGSHTWEHPNMTTIP
MUL_0184|M.ulcerans_Agy99 DNDAKATFFLIGNKV----AANPAAAKRIAEAGMEIGSHTWEHPNMTTIP
MAV_1218|M.avium_104 DNDAKATFFLIGNKV----AANPAGAKRIADAGMEIGNHTWEHPNMTTIP
Mb1126|M.bovis_AF2122/97 DNDAKATFFLIGNKV----AANPAGARRIADAGMEIGSHTWEHPNMTTIP
Rv1096|M.tuberculosis_H37Rv DNDAKATFFLIGNKV----AANPAGARRIADAGMEIGSHTWEHPNMTTIP
MLBr_01950|M.leprae_Br4923 DNDAKATFFLIGNKV----ATNPAGAKRIVDAGMEIGSHTWEHPNMTMLS
MAB_0459c|M.abscessus_ATCC_199 DRGAKATFFLIGNKV----AADPAAARRIAAAGMEVANHTWEHPNMATIP
Mflv_2063|M.gilvum_PYR-GCK AQGAKATFFVIGEKV----AADPAATRRITGAGMQVGNHTWQHLDMTTLA
Mvan_4651|M.vanbaalenii_PYR-1 ANNAEATFFLIGDKV----AADPAAARRIVDAGMEIGNHTWSHPDLTAIP
MSMEG_5962|M.smegmatis_MC2_155 ERGITAEFFLLAGELGKLGRVDDTGVRELLDAGMSIGSHGWAHRDWRRID
.. * **::. :: .: :..:.: ***.:..* * * : :
MMAR_4371|M.marinum_M EADIAGQFSRANDAIKAATGRTPTLYRPAGGLSNAAVRQTAEKFGQAEIL
MUL_0184|M.ulcerans_Agy99 EADIAGQFSRANDAIKAATGRTPTLYRPAGGLSNAAVRQTAAKYGQAEIL
MAV_1218|M.avium_104 TEDVAAQFTRANDAIHAATGRTPNLYRPAGGLSNPVVRQTAGQLGLAEIL
Mb1126|M.bovis_AF2122/97 PEDIPGQFSRANDVIAAATGRTPTLYRPAGGLSNDAVRQAAAKVGQAEIL
Rv1096|M.tuberculosis_H37Rv PEDIPGQFSRANDVIAAATGRTPTLYRPAGGLSNDAVRQAAAKVGQAEIL
MLBr_01950|M.leprae_Br4923 TEDIADQFSRANHAITVATGRTPTLWRPAGGLSNEAVRQVAGRYGQAEIL
MAB_0459c|M.abscessus_ATCC_199 TADIGAQLSKGVEAIAAATGIAPHLYRPAGGVSNAAVRAEAGRQHLAEIL
Mflv_2063|M.gilvum_PYR-GCK PPDVAAQFARATEAIAAATGQRTPLARTGFGAIDDSVLAEAGRQGLTAVN
Mvan_4651|M.vanbaalenii_PYR-1 PQELPAQLSRATEVLERATGERPTLMRPPFGAVDDAVLAAAGSQGLAVVN
MSMEG_5962|M.smegmatis_MC2_155 DAQAHEEFEDAHERLSAVTGSSVSRVAIPFGSYDRHVLRRLR--------
: :: . . : .** * : *
MMAR_4371|M.marinum_M WDVIPFDWANDSNTAATRYMLMRQIKPGSVVLFHDTYSSTVDLVYQFIPV
MUL_0184|M.ulcerans_Agy99 WDVIPFDWANDSNTAATRYMLMRQIKPGSVVLFHDTYSSTVDLVYQFIPV
MAV_1218|M.avium_104 WDVIPFDWANDSNTAATRYMLMTYIKPGSVVLFHNTYSSTVDLVYQFIPV
Mb1126|M.bovis_AF2122/97 WDVIPFDWINDSNTAATRHMLMTQIKPGSVVLFHDTYSSTVDVVYQFIPV
Rv1096|M.tuberculosis_H37Rv WDVIPFDWINDSNTAATRHMLMTQIKPGSVVLFHDTYSSTVDVVYQFIPV
MLBr_01950|M.leprae_Br4923 WDVIPFDWTNDSDTVATRCMLMMQIKPGSVVLFHDTYSSTVDLVYQFIPV
MAB_0459c|M.abscessus_ATCC_199 WDVIPFDWINDADTAATVYMLKTQIKPGSVVLLHDTYSSTVDIVYQFLPV
Mflv_2063|M.gilvum_PYR-GCK WDVNPRDWHHDADPDAIRDAVLTQVRPGAVVLLHDTVAATVGAMADVVPA
Mvan_4651|M.vanbaalenii_PYR-1 WDVIPLDWENDTDIAATRAALTEQIRPNSVVLLHDTFASTVDLMAEFVPV
MSMEG_5962|M.smegmatis_MC2_155 ------------DAGVTRVYTSDGGRARPEAWLQARTSLRHDIDADWIYR
: . :. . . :: : . : :
MMAR_4371|M.marinum_M LKANGYRLVTVSELLGPRAPGSSYGSRDNGPPVDELRDIPPSDIPPLPNT
MUL_0184|M.ulcerans_Agy99 LKANGYRLVTVSELLGPRAPGNSYGSRDNGPPVDELRDIPPSDIPPLPNT
MAV_1218|M.avium_104 LKANGYRLVTVSELLGPRAPGSSYGSRENGPPVNGLRDIPASDIPKLPDT
Mb1126|M.bovis_AF2122/97 LKANGYRLVTVSELLGPRAPGSSYGSRENGPPVNELRDIPASEIPPLPNT
Rv1096|M.tuberculosis_H37Rv LKANGYRLVTVSELLGPRAPGSSYGSRENGPPVNELRDIPASEIPPLPNT
MLBr_01950|M.leprae_Br4923 LKANGYRLVTVTELLGPRAPGSSYGRRENGPPVNQLHDIPANLIPALPNT
MAB_0459c|M.abscessus_ATCC_199 LIANGYHMVTVSHLLGDRAPGTSYGGRENGPPVNDIHDIPPAQIPVLPAT
Mflv_2063|M.gilvum_PYR-GCK LRARGYHLVTVSQLLGPLTPGSLYGSRERA--------------------
Mvan_4651|M.vanbaalenii_PYR-1 LRANGYHLVTVSQMIGPRPPGSLYGTPP----------------------
MSMEG_5962|M.smegmatis_MC2_155 LLDPHTPLVLSARNSAARAVKRWRG-------------------------
* :* :. . . *
MMAR_4371|M.marinum_M PSPKPMPNFPITDIAGQNSGGPNNGA
MUL_0184|M.ulcerans_Agy99 PSPKPMPNFPITDIAGQNSGGPNNGA
MAV_1218|M.avium_104 PSPKPMPNFPITDIPGQNSGGPNNGA
Mb1126|M.bovis_AF2122/97 SSPKPMSNFPITDIAGQNSGGPNNGA
Rv1096|M.tuberculosis_H37Rv SSPKPMPNFPITDIAGQNSGGPNNGA
MLBr_01950|M.leprae_Br4923 SSPKPMPNFPITDISGQNSGGSNDGA
MAB_0459c|M.abscessus_ATCC_199 PSPQPAPNLPITDIAGQNPGGPQ---
Mflv_2063|M.gilvum_PYR-GCK --------------------------
Mvan_4651|M.vanbaalenii_PYR-1 --------------------------
MSMEG_5962|M.smegmatis_MC2_155 --------------------------