For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSKTVEVAASAETITSIVSDFEAYPQWNPEIKGCWILARYNDGRPSQLRLDVEIQGQSGVFITAVYYPAE NQIFTMLQQGDHFTKQEQRFSIVPLGPDSTLLQVDLDVEVKLPVPGPMVKKLAGETLEHLAKALEGRVEQ LTQS
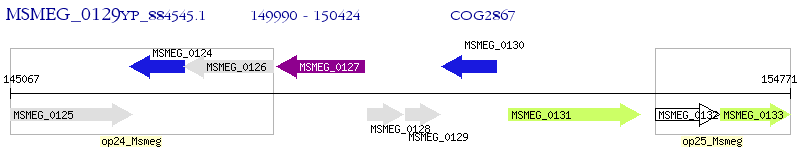
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0129 | - | - | 100% (144) | cyclase/dehydrase family protein |
| M. smegmatis MC2 155 | MSMEG_5736 | - | 2e-13 | 27.74% (137) | cyclase/dehydrase |
| M. smegmatis MC2 155 | MSMEG_4251 | - | 6e-06 | 20.86% (139) | cyclase/dehydrase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0169 | TB18.5 | 1e-44 | 59.03% (144) | hypothetical protein Mb0169 |
| M. gilvum PYR-GCK | Mflv_0716 | - | 6e-61 | 77.08% (144) | cyclase/dehydrase |
| M. tuberculosis H37Rv | Rv0164 | TB18.5 | 1e-44 | 59.03% (144) | hypothetical protein Rv0164 |
| M. leprae Br4923 | MLBr_02629 | - | 6e-46 | 63.12% (141) | hypothetical protein MLBr_02629 |
| M. abscessus ATCC 19977 | MAB_4572c | - | 8e-36 | 53.90% (141) | hypothetical protein MAB_4572c |
| M. marinum M | MMAR_0407 | - | 8e-44 | 60.56% (142) | hypothetical protein MMAR_0407 |
| M. avium 104 | MAV_5019 | - | 1e-43 | 58.87% (141) | cyclase/dehydrase superfamily protein |
| M. thermoresistible (build 8) | TH_0157 | - | 3e-59 | 75.89% (141) | CONSERVED HYPOTHETICAL PROTEIN TB18.5 |
| M. ulcerans Agy99 | MUL_1057 | - | 7e-44 | 61.27% (142) | hypothetical protein MUL_1057 |
| M. vanbaalenii PYR-1 | Mvan_0129 | - | 8e-56 | 70.83% (144) | cyclase/dehydrase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0169|M.bovis_AF2122/97 MTAISCSPRPRYASRMPVLSKTVEVTADAASIMAIVADIERYPEWNEGVK
Rv0164|M.tuberculosis_H37Rv MTAISCSPRPRYASRMPVLSKTVEVTADAASIMAIVADIERYPEWNEGVK
MMAR_0407|M.marinum_M --------MPRYASPMPVLSKTVEVSADAGSIMGIVADIERYPEWNEGVK
MUL_1057|M.ulcerans_Agy99 --------MPRYASPMPVLSKTVEVSADAGSIMGIVADIERYPEWNEGVK
MAV_5019|M.avium_104 ---------------MPVLSKTVEVNTDAAAIMAIVADFERYPEWSDGVT
MLBr_02629|M.leprae_Br4923 -----MGSVPGYASPMPVMSKTVEVRATAASIMAIVTDFEAYPQWNDGVK
Mflv_0716|M.gilvum_PYR-GCK ---------------MPLVSKTVEVSASAETIMAIVADFEKYPEWNEEIK
Mvan_0129|M.vanbaalenii_PYR-1 ---------------MPLVSKTVEVEAAAETIMGIVADFESYPQWNEEIK
TH_0157|M.thermoresistible__bu ------------------VSKTVEVAAPADRIMAIVADFESYPLWNQEIK
MSMEG_0129|M.smegmatis_MC2_155 ------------------MSKTVEVAASAETITSIVSDFEAYPQWNPEIK
MAB_4572c|M.abscessus_ATCC_199 ---------------MPVVSQTVEVAAPPQVIVSIVTNYEAYPEWNKEIA
:*:**** : . * .**:: * ** *. :
Mb0169|M.bovis_AF2122/97 GAWVLARYDDGRPSQVRLDTAVQGIEGTYIHAVYYPGENQIQTVMQQGEL
Rv0164|M.tuberculosis_H37Rv GAWVLARYDDGRPSQVRLDTAVQGIEGTYIHAVYYPGENQIQTVMQQGEL
MMAR_0407|M.marinum_M GAWVLARYDDGRPSQVRLDASVQGFEGVYIHAVYYPGENQIQTVMQQGDL
MUL_1057|M.ulcerans_Agy99 GAWVLARYDDGRPSQVRLDASVQGFEGVYIHAVYYPGENQIQTVMQQGDL
MAV_5019|M.avium_104 GCWVLARYDDGRPSQLRLDAAYQGFEGVYIQAVYYPGPNQIQTVMQQGEL
MLBr_02629|M.leprae_Br4923 GVWVLARYDDGRPSQLRLDTEIQGTKCTYIQAVYYPATNQIQTIMQQGDL
Mflv_0716|M.gilvum_PYR-GCK GCWILARYNDGRPSQLRLDVVVQGQAGTFITAVYYPAENQIFTMLQQGDH
Mvan_0129|M.vanbaalenii_PYR-1 GCWVLARYNDGRPSQLRLDVVVQGQAGTFITAVYYPGENQIYTVLQQGDH
TH_0157|M.thermoresistible__bu GCWVLARYNDGRPSQLRLDVVVQGQSGTFITAVYYPAPNQIFTMLQQGDH
MSMEG_0129|M.smegmatis_MC2_155 GCWILARYNDGRPSQLRLDVEIQGQSGVFITAVYYPAENQIFTMLQQGDH
MAB_4572c|M.abscessus_ATCC_199 SVDILQRLPDGRPHIVRLKVETSGMSSTNVAEIAYLNAAQVATRLLESDI
. :* * **** :**.. .* . : : * *: * : :.:
Mb0169|M.bovis_AF2122/97 FAKQEQLFSVVATGA-ASLLTVDMDVQVTMPVPEPMVKMLLNNVLEHLAE
Rv0164|M.tuberculosis_H37Rv FAKQEQLFSVVATGA-ASLLTVDMDVQVTMPVPEPMVKMLLNNVLEHLAE
MMAR_0407|M.marinum_M FLKQEQLFSVVEAGA-ASLLTVDIDVEPSMPVPPPMVKLLLNNVLEQLAA
MUL_1057|M.ulcerans_Agy99 FLKQEQLFSVVEAGA-VSLLTVDIDVEPSMPVPPPMVKLLLNNVLEQLAA
MAV_5019|M.avium_104 FKKQEQLFSVVEMGA-SSLLTVDIDVEPSMPVPAPMVKSMLNNVLDHLAD
MLBr_02629|M.leprae_Br4923 FTKQEQLFSAVEIGA-ASLLTVDIDVESSMPVPAPMVKALLNNVLDNLAE
Mflv_0716|M.gilvum_PYR-GCK FTKQEQRFAVVPMGP-TSLLTVDLDVEVSMPVPAVMVKKVIGDTLDYLAE
Mvan_0129|M.vanbaalenii_PYR-1 LSKQEQRFSVVAMGA-TSLLTVDLEVEVKMAVPNAMVKKIVGDTLDYLAD
TH_0157|M.thermoresistible__bu FEKQEQTFSVVAMGPNSSLLTVDLDVETKMPVPKSMVKKLAGDTLDYLAD
MSMEG_0129|M.smegmatis_MC2_155 FTKQEQRFSIVPLGPDSTLLQVDLDVEVKLPVPGPMVKKLAGETLEHLAK
MAB_4572c|M.abscessus_ATCC_199 FEKQEQTFSIVPMGQ-TCLLTVDMDVETKLPIPKPMVKKLANQVLEHLAE
: **** *: * * ** **::*: .:.:* *** : .:.*: **
Mb0169|M.bovis_AF2122/97 NLKQRAEQLAAS--------------
Rv0164|M.tuberculosis_H37Rv NLKQRAEQLAAS--------------
MMAR_0407|M.marinum_M NLKLRAEQLTAG--------------
MUL_1057|M.ulcerans_Agy99 NLKLRAEQLTAG--------------
MAV_5019|M.avium_104 NLKQRAEQLAAN--------------
MLBr_02629|M.leprae_Br4923 NLKLRAEQLAAN--------------
Mflv_0716|M.gilvum_PYR-GCK NLKTRAEHLTAS--------------
Mvan_0129|M.vanbaalenii_PYR-1 NLKTRAEQLAST--------------
TH_0157|M.thermoresistible__bu NLKTRAEQLAS---------------
MSMEG_0129|M.smegmatis_MC2_155 ALEGRVEQLTQS--------------
MAB_4572c|M.abscessus_ATCC_199 GLKGRAEAIASGQLQPAPVQQPPTQA
*: *.* ::