For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PVARIGADHLGHSRISMTQDKHMARGRVHTVVADLLDRTINDE*
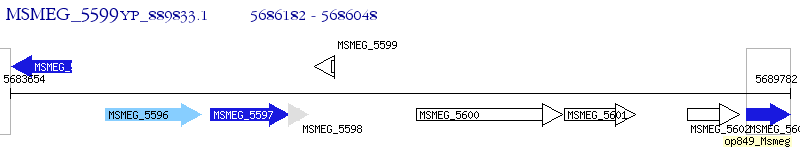
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5599 | - | - | 100% (44) | hypothetical protein MSMEG_5599 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1816c | - | 6e-13 | 70.73% (41) | Phage integrase |
| M. marinum M | MMAR_3607 | - | 6e-13 | 74.42% (43) | phage integrase |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5599|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1816c|M.abscessus_ATCC_199 -----------------------------MAGRPPLRIGQHGKISRQYVG
MMAR_3607|M.marinum_M MQSGDSQRWNNPWPGKSWPGMSSLAEVGTMAGRPQLRIGTHGQIKRIYTG
MSMEG_5599|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1816c|M.abscessus_ATCC_199 GGVWVARCRYRDSDGVTRIVERRGPVDDFDKHGKLAEDALIEALSDRRLP
MMAR_3607|M.marinum_M GGVWLARCRYRDTDGVTRIVQRLGPADEYDQHGKLAADALIAALAQRRHS
MSMEG_5599|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1816c|M.abscessus_ATCC_199 ADTDTIGYDTLVMALVDQHIGRLSDDGRSPATVATYKFASDKLRNFLGGV
MMAR_3607|M.marinum_M ANHGEIGPDTKVTDLVEQHLSRLAEDGRSPVTMSTYRFTTKNLKKFIGGL
MSMEG_5599|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1816c|M.abscessus_ATCC_199 RVSEAHAARMDAALRSMRTAHGTTMARQAKTILQGGLQLAVMANVIPANP
MMAR_3607|M.marinum_M RVSEATPARMDATLRSMRSAHGATMARQSKSILRGALQLAVMASVLGANP
MSMEG_5599|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1816c|M.abscessus_ATCC_199 VRDVQSLRSKTRPKGATALSADQLRTLLAEIRASEYCAGNDLADPLIILM
MMAR_3607|M.marinum_M VRDVQPLRSQSQPKGAVALTADQLRELLARLQTSDFCKEHDLIDPITLLI
MSMEG_5599|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1816c|M.abscessus_ATCC_199 ATGLRRSELLALRWSDFDANAGTLTVTGKLVRATGAGLQRIDEAKTAAGM
MMAR_3607|M.marinum_M ATGLRRSELLGLRWVDFDEKASMLAITGKVIRVPGEGLLRVDETKSAAGR
MSMEG_5599|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1816c|M.abscessus_ATCC_199 RTVALPSFAVTALRHRQGRDYVGQQLMIFPSTAGTWRDPNNFGKQWRAVR
MMAR_3607|M.marinum_M RTIPLPKFAVDVLGKRRQLPYLGEQVVIFPSTAGTLRDPNNFGKEWRTAR
MSMEG_5599|M.smegmatis_MC2_155 ----MP--------------------VARIGADHLGHSRISMTQDKHMAR
MAB_1816c|M.abscessus_ATCC_199 DDLGVPGVTTHSFRKTMATLIDDNGLSARVGADQLGHAKVSMTQDRYMAR
MMAR_3607|M.marinum_M EELGVAEVTTHSFRKTVATLIDDEGLSARIGADHLGHSHVSMTQDRYMTR
:. **:***:***:::*****::*:*
MSMEG_5599|M.smegmatis_MC2_155 GRVHTVVADLLDRT--INDE
MAB_1816c|M.abscessus_ATCC_199 GKVHHEVADLLDAT--ISDE
MMAR_3607|M.marinum_M GRIHTQVAHLLDRAVRINDE
*::* **.*** : *.**