For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPLASGQVFAGFTIVRLVGTGGMGEVYLAKHPRLPREDALKVLPISVSADDEFRQRFIREADMAATLWHP HIVGVHDRGEFEGRLWISMDYVDGHDAARLLHDKYPKGMPADDVVEIVTAVGDALDYAHQRQLLHRDVKP ANILVTEKHGTKRRIMLTDFGIARRTDDVNGLTSTNITVGSMSYTSPEQLMGQPLDGRADQYSLAATAYR LLTGAPPFSHSNPAVVISHHLNSPPPKLGDTKPALAKLDSVMAKALAKDAKDRFDTCHDFAVALAEANKG DESAAPVTAAPPPVITPPVQTAPPTTPLKAQTLPAPPTPPTPLSPSNPQAPTASQPPVFTSSWAGHDDPS GRQPVAVPAPAPPPPPPPSNFGPPPPAYSPPPRKSGGDRRKLIIAAAAIGAVVLLVGGITLAVTSGGDND SAGAAATTSETASETPVTTTPKKPGEHAYTIADYIRDNKISETPVHRGDPNTPELTMPTPPGWADAGTRT PAWAYSAIVNDPASPTDPPSVISLISKLDGNVDPEKLLEFAPNELQNLTEWEPIGGVTRSNLSGFPSVQL GGSYLKDGKRRAITQKTVVVDIPSGIYVLQINADSLYRDFGALVDVNKAIDKEATIKP
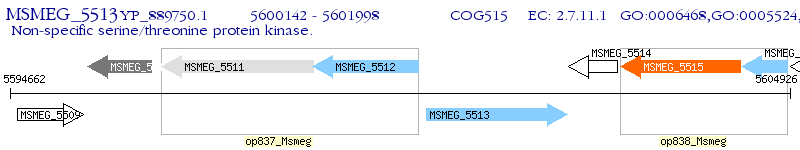
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5513 | - | - | 100% (618) | serine/threonine-protein kinase PknE |
| M. smegmatis MC2 155 | MSMEG_0886 | - | e-118 | 61.31% (336) | serine/threonine-protein kinase PknD |
| M. smegmatis MC2 155 | MSMEG_3677 | - | e-107 | 56.13% (367) | serine/threonine protein kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1775 | pknF | 1e-110 | 49.33% (450) | anchored-membrane serine/threonine-protein kinase PKNF |
| M. gilvum PYR-GCK | Mflv_3427 | - | 1e-112 | 52.93% (410) | protein kinase |
| M. tuberculosis H37Rv | Rv1746 | pknF | 1e-110 | 63.67% (300) | anchored-membrane serine/threonine-protein kinase PKNF |
| M. leprae Br4923 | MLBr_00016 | pknB | 2e-48 | 40.36% (275) | putative serine/threonine protein kinase |
| M. abscessus ATCC 19977 | MAB_4435 | - | 6e-66 | 43.20% (331) | serine/threonine-protein kinase PknJ |
| M. marinum M | MMAR_4174 | pknF | 1e-110 | 55.64% (381) | anchored-membrane serine/threonine-protein kinase PknF |
| M. avium 104 | MAV_3145 | - | 1e-102 | 51.47% (373) | PknF protein |
| M. thermoresistible (build 8) | TH_1607 | pknI | 1e-86 | 66.37% (226) | PROBABLE TRANSMEMBRANE SERINE/THREONINE-PROTEIN KINASE I |
| M. ulcerans Agy99 | MUL_2200 | - | 1e-109 | 56.29% (350) | membrane-anchored serine/threonine-protein kinase |
| M. vanbaalenii PYR-1 | Mvan_5635 | - | 1e-105 | 61.20% (317) | protein kinase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5513|M.smegmatis_MC2_155 --------------------------------------------------
TH_1607|M.thermoresistible__bu --------------------------------------------------
Mb1775|M.bovis_AF2122/97 --------------------------------------------------
Rv1746|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_3427|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5635|M.vanbaalenii_PYR-1 --------------------------------------------------
MUL_2200|M.ulcerans_Agy99 --------------------------------------------------
MMAR_4174|M.marinum_M --------------------------------------------------
MAV_3145|M.avium_104 MNEHKGTTRRIGAGRRLLSNPRQARAALRAGFQSLPGMPIGQLFRRIPPV
MAB_4435|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00016|M.leprae_Br4923 --------------------------------------------------
MSMEG_5513|M.smegmatis_MC2_155 ---------------------MPLASGQVFAGFTIVRLVGTGGMGEVYLA
TH_1607|M.thermoresistible__bu ---------------------MPLADGQQFAEYTILRRLGAGGMGEVYLA
Mb1775|M.bovis_AF2122/97 ---------------------MPLAEGSTFAGFTIVRQLGSGGMGEVYLA
Rv1746|M.tuberculosis_H37Rv ---------------------MPLAEGSTFAGFTIVRQLGSGGMGEVYLA
Mflv_3427|M.gilvum_PYR-GCK ---------------------MPLAAGETFAGYTIVRLLGSGGMGEVYLA
Mvan_5635|M.vanbaalenii_PYR-1 ---------------------MPLSDGEVIAGYTIVRMLGSGGMGKVYLA
MUL_2200|M.ulcerans_Agy99 ---------------------MPLNEGDIFAGYVIQRLLGTGGMGEVYLA
MMAR_4174|M.marinum_M ---------------------MPLPPGSAFAGYTIVGLLGSGAMGEVYLA
MAV_3145|M.avium_104 DSRPHPEPAAAQGNSEPGGPRLTIGNGASFAGYTILRQLGAGGMAEVYLA
MAB_4435|M.abscessus_ATCC_1997 -----MTSSDRSSPGPGGQQPGDLPPGTVVSGYQIERVLGRGGMGTVYLA
MLBr_00016|M.leprae_Br4923 ----------------------MTTPPHLSDRYELGDILGFGGMSEVHLA
: : :* *.*. *:**
MSMEG_5513|M.smegmatis_MC2_155 KHPRLPREDALKVLPISVSADDEFRQRFIREADMAATLWHPHIVGVHDRG
TH_1607|M.thermoresistible__bu RHPRLPRNDALKVLSAEVSADDEYRRRFEQEADIAATLWHPHIVAVHDRG
Mb1775|M.bovis_AF2122/97 RHPRLPRQDALKVLRADVSADGEYRARFNREADAAASLWHPHIVAVHDRG
Rv1746|M.tuberculosis_H37Rv RHPRLPRQDALKVLRADVSADGEYRARFNREADAAASLWHPHIVAVHDRG
Mflv_3427|M.gilvum_PYR-GCK QHPRLPRRDALKVLPAAVSADVEYRKRFEREADIAATLWHPHIVGVHDRG
Mvan_5635|M.vanbaalenii_PYR-1 KHPRLPRYDALKVLSTTVCADSEYRERFHREADIAATLWHPHIVAVHDRG
MUL_2200|M.ulcerans_Agy99 QHPRLPRLDALKILSLDATDDDEFRARFTREAELAATLWHPHIVGVHDRG
MMAR_4174|M.marinum_M QHPRLPRRDALKVLPPDVSADTAFRERFQREADLAATLFHPHIVGVHDRG
MAV_3145|M.avium_104 LHPRLPRRDVIKVLAEAVTVDPEFRERFNREADLAATLWHPHIVGVHDRG
MAB_4435|M.abscessus_ATCC_1997 ANPNLQRSEALKILSAQSSRDPEFRARFMREASLAASLDHPNIVTVHNRG
MLBr_00016|M.leprae_Br4923 RDIRLHRDVAVKVLRADLARDPSFYLRFRREAQNAAALNHPSIVAVYDTG
. .* * .:*:* * : ** :**. **:* ** ** *:: *
MSMEG_5513|M.smegmatis_MC2_155 EF----EGRLWISMDYVDGHDAARLLHDKYPKGMPADDVVEIVTAVGDAL
TH_1607|M.thermoresistible__bu EY----DGQLWIAMDHVDGTDVGRLLGEQYPNGMPPDAVIRIITAVADAL
Mb1775|M.bovis_AF2122/97 EF----DGQLWIDMDFVDGTDTVSLLRDRYPNGMPGPEVTEIITAVAEAL
Rv1746|M.tuberculosis_H37Rv EF----DGQLWIDMDFVDGTDTVSLLRDRYPNGMPGPEVTEIITAVAEAL
Mflv_3427|M.gilvum_PYR-GCK ED----AGRIWISMDYVEGADAAHLLAEDYRTGLPVAEVAQIVTAVADAL
Mvan_5635|M.vanbaalenii_PYR-1 EY----EGRLWISMDHVEGTDAAWLLAERYPHGMPPALVIRIVTAVGEAL
MUL_2200|M.ulcerans_Agy99 EF----DGRLWISMDYVDGTDTRHLVEQRYRSGMPLEDVIEIVTAVAEAL
MMAR_4174|M.marinum_M EF----NGQLWISMDYIEGTDAAALIRSQYPAGMPVAEVVAVVTAVAGAL
MAV_3145|M.avium_104 EF----NGHLWISMDYVEGTDASRLVKESYADGMPLDEVSAIVQAVAGAL
MAB_4435|M.abscessus_ATCC_1997 ETP---EGDLWIAMQYVQGSDAH---SESASGRMSLTRAVHIISEVAKAL
MLBr_00016|M.leprae_Br4923 EAETSAGPLPYIVMEYVDGATLRDIVHTDGP--MPPQQAIEIVADACQAL
* :* *:.::* :. . :: . **
MSMEG_5513|M.smegmatis_MC2_155 DYAHQRQLLHRDVKPANILVTEKHGTK--RRIMLTDFGIARRTDDVNGLT
TH_1607|M.thermoresistible__bu DHAHSRNLLHRDVKPANILLAAGDGEQGAERVLLADFGIARWIDRSSGLT
Mb1775|M.bovis_AF2122/97 DYAHERRLLHRDVKPANILIANPDSPD--RRIMLADFGIAGWVDDPSGLT
Rv1746|M.tuberculosis_H37Rv DYAHERRLLHRDVKPANILIANPDSPD--RRIMLADFGIAGWVDDPSGLT
Mflv_3427|M.gilvum_PYR-GCK DYAHDRHLLHRDVKPANILIARPDSNT--RRIMLADFGIARWDNDISGLT
Mvan_5635|M.vanbaalenii_PYR-1 DYAHQRGLLHRDVKPANILIADPETEN--ERIMLADFGIARRVGEVSTLT
MUL_2200|M.ulcerans_Agy99 DFAHERRLLHRDVKPANILVTEPSESAR-RRVLLTDFGIAREIDDKHRLT
MMAR_4174|M.marinum_M DYAHQRGLLHRDVKPANILLTGPDNAGG-PRIFLADFGIARQLDDISGIT
MAV_3145|M.avium_104 DYAHARGLLHRDVKPANILLTHP-EAGE-RRILLADFGVARHLGDISGIT
MAB_4435|M.abscessus_ATCC_1997 DYLHARGLVHRDVKPANILLSAGD-----RRIMLGDFGLTRALDDSNSVT
MLBr_00016|M.leprae_Br4923 NFSHQNGIIHRDVKPANIMISATN------AVKVMDFGIARAIADSTSVT
:. * . ::*********::: : : ***:: :*
MSMEG_5513|M.smegmatis_MC2_155 STNITVGSMSYTSPEQLMG-QPLDGRADQYSLAATAYRLLTGAPPFSHSN
TH_1607|M.thermoresistible__bu ETDMTVGTVAYAAPEQLRA-ADIDGRADQYALAATAFQLMTGAPPFQHSN
Mb1775|M.bovis_AF2122/97 ATNMTVGTVSYAAPEQLMG-NELDGRADQYALAATAFHLLTGSPPFQHAN
Rv1746|M.tuberculosis_H37Rv ATNMTVGTVSYAAPEQLMG-NELDGRADQYALAATAFHLLTGSPPFQHAN
Mflv_3427|M.gilvum_PYR-GCK ATNMTVGTVSYAAPEQLMG-QDLDGRADQYALAATAFHLLTGSPPFSHSN
Mvan_5635|M.vanbaalenii_PYR-1 GTSMTVGTVAYSAPEQLTADEHIDGRADQYALAATAFQLLTGSPPFQHSN
MUL_2200|M.ulcerans_Agy99 EAQMAIGTVAYAAPEQLTG-RPLDGRADQYALAATAFHLLAGVPPFDHSN
MMAR_4174|M.marinum_M ATNLTVGTAAYAAPEQLTG-ADVDGRADQYALAATAFHLLTGAPPYQHAN
MAV_3145|M.avium_104 ETNVAVGTVAYAAPEQLTG-SPIDGRADQYALAATAFHLLTGAPPFQHSN
MAB_4435|M.abscessus_ATCC_1997 SAGNVTATIAYAAPEILSG-TAVDGRADVYALGCTLFRMLTGQPPFGSGN
MLBr_00016|M.leprae_Br4923 QTAAVIGTAQYLSPEQARG-DSVDARSDVYSLGCVLYEILTGEPPFIGDS
: . .: * :** . :*.*:* *:*... :.:::* **: .
MSMEG_5513|M.smegmatis_MC2_155 -PAVVISHHLNSPPPKLGDTKP-ALAKLDSVMAKALAKDAKDRFDTCHDF
TH_1607|M.thermoresistible__bu -PAXXXCGR-----------------------------------------
Mb1775|M.bovis_AF2122/97 -PAVVISQHLSASPPAIGDRVP-ELTPLDPVFAKALAKQPKDRYQRCVDF
Rv1746|M.tuberculosis_H37Rv -PAVVISQHLSASPPAIGDRVP-ELTPLDPVFAKALAKQPKDRYQRCVDF
Mflv_3427|M.gilvum_PYR-GCK -PAVVISRHLNSAPATVAAHRP-ELAAVDPVLTRALAKNPADRYPRCADF
Mvan_5635|M.vanbaalenii_PYR-1 -PAIVISQHLTAQPPSISVHRP-ELSSLGMAFEKALAKSPADRFDRCVDF
MUL_2200|M.ulcerans_Agy99 -RAVVVGQHLNMPPPRISKRHP-KLAHLDAAFAKALAKDPDDRYPRCIDF
MMAR_4174|M.marinum_M -SVAVISQHLTAPPPALGDRRP-ELAPLDRVFAAALAKQPNERYWRCADF
MAV_3145|M.avium_104 -PIAVIGQHLHEDPPRLSDFRP-ELAALDEVFCQALAKVPEDRFDRCRAF
MAB_4435|M.abscessus_ATCC_1997 SLPAIINAHLSAPPPRISQLAP-VPESWDALIAKAMAKTPADRYQSAGEL
MLBr_00016|M.leprae_Br4923 -PVSVAYQHVREDPIPPSQRHEGISVDLDAVVLKALAKNPENRYQTAAEM
:
MSMEG_5513|M.smegmatis_MC2_155 AVALAE-----------ANKGDESAAPVTAAPPPVITPPVQTAPPTTPLK
TH_1607|M.thermoresistible__bu --------------------------------------------------
Mb1775|M.bovis_AF2122/97 ARALGH-----------RLGGAGDPDDTRVSQPVAVAAPAKRS-------
Rv1746|M.tuberculosis_H37Rv ARALGH-----------RLGGAGDPDDTRVSQPVAVAAPAKRS-------
Mflv_3427|M.gilvum_PYR-GCK AREFNA-----------RLDAG---------------APAHTS-------
Mvan_5635|M.vanbaalenii_PYR-1 ARALAN-----------RSGTELPGSAFDPDATRHAAAPTGPR-------
MUL_2200|M.ulcerans_Agy99 AKALAAGPGSIAEHQARRGATSTEPFDVNAEMSAEALEPARPSGTAPSAQ
MMAR_4174|M.marinum_M AQALS----------------QVASPQG-------AAAAVSP--SDPTLP
MAV_3145|M.avium_104 AAAVSRECDGAAAIGPDARSRTVASPAHRRRGPGRVIAAVTHRFSPQTRW
MAB_4435|M.abscessus_ATCC_1997 AMAAQQILTDP----AQHRQTVTQPVPATTPAPIAAPEPVHAMAPLAMAD
MLBr_00016|M.leprae_Br4923 RADLIRVHSGQPPEAPKVLTDADRSCLLSSGAGNFGVPRTDALSRQSLDE
MSMEG_5513|M.smegmatis_MC2_155 AQTLPAPPTPPTPLSPSNPQAPTASQPPVFTSSWAGHDDPSGRQPVAVPA
TH_1607|M.thermoresistible__bu --------------------------------------------------
Mb1775|M.bovis_AF2122/97 -------------------LLRTAVIVPAV--------------------
Rv1746|M.tuberculosis_H37Rv -------------------LLRTAVIVPAV--------------------
Mflv_3427|M.gilvum_PYR-GCK -------------------RIRPAVLIPVV--------------------
Mvan_5635|M.vanbaalenii_PYR-1 -------------------HAKTAAAPRSR--------------------
MUL_2200|M.ulcerans_Agy99 AQPHPSEAGVSTASLGAADQPGAGKARHTKRLERMTVDGTAVSVQKRPAG
MMAR_4174|M.marinum_M AAISVPPEYAAATQAAPAPMPWYPTAP-----------------------
MAV_3145|M.avium_104 AAALVCAVLVAVAATWSVLYSFQPGAPPAN--------------------
MAB_4435|M.abscessus_ATCC_1997 TGQSMLKHKRIAAIIGVMVLVVASSVVAVLIWPRADEG------------
MLBr_00016|M.leprae_Br4923 TESDGSIGRWVAVVAVLAVLTIAIVAAFNTFGGNTRDVQVPDVRG-----
MSMEG_5513|M.smegmatis_MC2_155 PAPPPPPPPSNFGPPPPAYSPPPRKSGGDRRKLIIAAAAIGAVV------
TH_1607|M.thermoresistible__bu --------------------------------------------------
Mb1775|M.bovis_AF2122/97 -----------------------------LAMLLVMAVAVTVRE------
Rv1746|M.tuberculosis_H37Rv -----------------------------LAMLLVMAVAVAVRE------
Mflv_3427|M.gilvum_PYR-GCK -----------------------------LAVLLVAAVAVAAFE------
Mvan_5635|M.vanbaalenii_PYR-1 -----------------------------RRVVLAGAGLVLALI------
MUL_2200|M.ulcerans_Agy99 PEQDEEVWLFCVQRYESTGEPHTVIPVELRGTSITGQLANGDAVGVSGIW
MMAR_4174|M.marinum_M --------------------------APARPRRQVSTIVVPVVLA-----
MAV_3145|M.avium_104 -------------------------PALASKPSPPAAVAAPIAGG-----
MAB_4435|M.abscessus_ATCC_1997 -------------------------TDPRTKASPSAAQVAPGASLAGQPN
MLBr_00016|M.leprae_Br4923 ------------QVSADAISALQNRGFKTRTLQKPDSTIPPDHVISTEPG
MSMEG_5513|M.smegmatis_MC2_155 ---LLVGGITLAVTSGGDNDS-------------------------AGAA
TH_1607|M.thermoresistible__bu --------------------------------------------------
Mb1775|M.bovis_AF2122/97 ---FQRADDERAAQPARTRTT-------------------------TSAG
Rv1746|M.tuberculosis_H37Rv ---FQRADDERAAQPARTRTT-------------------------TSAG
Mflv_3427|M.gilvum_PYR-GCK ---FLRAD--RADRADRADPP-------------------------PMAR
Mvan_5635|M.vanbaalenii_PYR-1 ---AVAAA-VFLARFERDVGR-------------------------GEAR
MUL_2200|M.ulcerans_Agy99 DGSTLFADAVVNYTAANRGRR-------------------------RSSA
MMAR_4174|M.marinum_M --LLLVSAVAFAVTQVLR--------------------------------
MAV_3145|M.avium_104 --PVLNGTYKLDYDQTKRTTNGIGIRHDGAGTNWWAFRSACTSSGCAATG
MAB_4435|M.abscessus_ATCC_1997 TVLDTLLPGQFDYPEGWEKNP---------------------------SP
MLBr_00016|M.leprae_Br4923 ANASVGAGDEITINVSTGPEQ-------------------------REVP
MSMEG_5513|M.smegmatis_MC2_155 ATTSET-------ASETPVTTTPKKPGEHAYTIADYIRDNKISETPVHRG
TH_1607|M.thermoresistible__bu --------------------------------------------------
Mb1775|M.bovis_AF2122/97 TTTSVAPASTTRPAPTTPTTTGAADTATASPTAAVVAIG-----------
Rv1746|M.tuberculosis_H37Rv TTTSVAPASTTRPAPTTPTTTGAADTATASPTAAVVAIG-----------
Mflv_3427|M.gilvum_PYR-GCK TTTITT-------SPTTSTTT-----ATPPATPASVVIG-----------
Mvan_5635|M.vanbaalenii_PYR-1 ATASAT-------ASTAPTGS-------GSVTLPVVVVG-----------
MUL_2200|M.ulcerans_Agy99 LKVEGGLKEGEASAPARPPGSKSRKGRAFAIVAALLALAAIAVAAVLTHG
MMAR_4174|M.marinum_M -------------PDPRQTTAAPAEAPQWQPYVDH---------------
MAV_3145|M.avium_104 TRLDDATHQAAGGPDGDQTDTLRFVGGYWQGAPEQQRVGCT---------
MAB_4435|M.abscessus_ATCC_1997 TFRSGEFNPAVPPGSATNIGGTPVGCNTIDPAWALRAKG-----------
MLBr_00016|M.leprae_Br4923 DVSSLNYTDAVKKLTSSGFKSFKQANSPSTPELLGKVIGTN---------
MSMEG_5513|M.smegmatis_MC2_155 D------PNTPELTMPTPPGWADAGTRTPAWAYSAIVND---PASPTDPP
TH_1607|M.thermoresistible__bu --------------------------------------------------
Mb1775|M.bovis_AF2122/97 --------------ALCFPLGSTGTTKTGATAYCSTLQG---TNTT----
Rv1746|M.tuberculosis_H37Rv --------------ALCFPLGSTGTTKTGATAYCSTLQG---TNTT----
Mflv_3427|M.gilvum_PYR-GCK --------------ANCAPLDATATTGDGTTAYCAAVPG---SAGT----
Mvan_5635|M.vanbaalenii_PYR-1 --------------ADCATLGAAGVTEAGDPAYCARLAS---SGAP----
MUL_2200|M.ulcerans_Agy99 FGLWSTSEPGPVVKAQQATVFSPGGTPDHPGQAGLAIDGDPNSAWPTDTY
MMAR_4174|M.marinum_M --------------AKQFAISLTSVNPQT---------------------
MAV_3145|M.avium_104 -------RPGGPAGATQQETIARSLAPQSDGTLRGTETETV-LSNECGAQ
MAB_4435|M.abscessus_ATCC_1997 --------------RETSTLYLRDRKKPEREILIRVVPAADALSTDG---
MLBr_00016|M.leprae_Br4923 -------PSANQTSAITNVITIIVGSGPETKQIPDVTGQIVEIAQKNLNV
MSMEG_5513|M.smegmatis_MC2_155 SVISLISKLDGNVDPEKLLEFAPNELQNLTEWEPIGGVTRSNLSGFPSVQ
TH_1607|M.thermoresistible__bu --------------------------------------------------
Mb1775|M.bovis_AF2122/97 ----IWSLTEDTVASPTVTATADPTEAPLPIEQESPIRVCMQQTGQTRRE
Rv1746|M.tuberculosis_H37Rv ----IWSLTEDTVASPTVTATADPTEAPLPIEQESPIRVCMQQTGQTRRE
Mflv_3427|M.gilvum_PYR-GCK ----IWSLTQGTLPAPTSTPPPDPTEQPLSDAEAFPVLTCMEQTGQTRRE
Mvan_5635|M.vanbaalenii_PYR-1 ----LWSLYPGEIPHPTGTSAPEGESQ----GPDTPVLVCMQQTGQSQVD
MUL_2200|M.ulcerans_Agy99 VDATPFPAFKQGVGLILRLAKPTALSEVTIDVPSTGTEVQIRAAGSATPA
MMAR_4174|M.marinum_M -----IDADIQRILDGSTGAFHSDFADKSAQFKQSIVDAGSATQGKATGA
MAV_3145|M.avium_104 GAVVRVPVVATRVGDVPPGVTVADPASVINASPTATAPAPPVLGGLCSDV
MAB_4435|M.abscessus_ATCC_1997 -----ASDALKRCDAVAYIIPGSSTTWRCSFEFDEPNPVQNGQKELTTTE
MLBr_00016|M.leprae_Br4923 YGFTKFSQASVDSPRPTGEVIGTNPPKDATVPVDSVIELQVSKGNQFVMP
MSMEG_5513|M.smegmatis_MC2_155 LGGSYLKDGKRRAITQKTVVVDIPSGIYVLQIN---ADSLYRDFGALVDV
TH_1607|M.thermoresistible__bu --------------------------------------------------
Mb1775|M.bovis_AF2122/97 CREEIRRSNGWP--------------------------------------
Rv1746|M.tuberculosis_H37Rv CREEIRRSNGWP--------------------------------------
Mflv_3427|M.gilvum_PYR-GCK CRRAIRESNGQPPLP-----------------------------------
Mvan_5635|M.vanbaalenii_PYR-1 CHDDILQENTDPGANDTQTPG-----------------------------
MUL_2200|M.ulcerans_Agy99 SLSDTTALTPNVALHPGHNRIRVDS--TTKTAN---VLVWISKLGTTKGE
MMAR_4174|M.marinum_M GLESISGTTAHVVVSVTSNITKS---TGVQQE----PRQWRLRIQIEKID
MAV_3145|M.avium_104 GKVAYDPTNNEQIVCEGSSWAKAPITMGVHAAGSSCDRPGTSVFAMSTSS
MAB_4435|M.abscessus_ATCC_1997 SCYMFPSGANSIRQHVSFNVVRGLLVYILASGASNRNDAAGSLSATALSF
MLBr_00016|M.leprae_Br4923 DLSGMFWADAEPRLRALGWTGVLDKGPDVDAGGSQHNRVAYQNPPAGAGV
MSMEG_5513|M.smegmatis_MC2_155 NKAIDKEATIKP-------
TH_1607|M.thermoresistible__bu -------------------
Mb1775|M.bovis_AF2122/97 -------------------
Rv1746|M.tuberculosis_H37Rv -------------------
Mflv_3427|M.gilvum_PYR-GCK -------------------
Mvan_5635|M.vanbaalenii_PYR-1 -------------------
MUL_2200|M.ulcerans_Agy99 SRNAISDITLRAR------
MMAR_4174|M.marinum_M DAYLASRVEFVT-------
MAV_3145|M.avium_104 DGYLLQCDPVTRTWTRPAG
MAB_4435|M.abscessus_ATCC_1997 ATTMKILRYGR--------
MLBr_00016|M.leprae_Br4923 NRDGIITLKFGQ-------