For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SIPHTAVVPAAGLGTRFLPATKTVPKELLPVVDTPGIELVAAEAAEAGAERLVIVTSEGKDGVVAHFVQD LVLEGTLEARGKKTMLEKVRRAPALIKVESVVQAEPLGLGHAVSCVESVLAADEDAISVLLPDDLVLPTG VLETMSKVRAKRGGSVLCAIEVPGDKISAYGVFDVETVPDAANPNVLRVKGMVEKPRPEDAPSHFAAAGR YVLDRAIFDALRRVPRGTGGEIQLTDAIALLIEEDHPVHVVVHRGARHDLGNPGGYLKAAVDFALERDDY GPELRQWLVERLGLAEK*
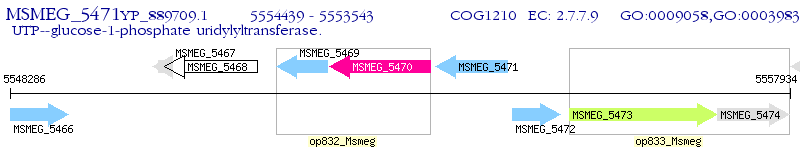
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5471 | galU | - | 100% (298) | UTP-glucose-1-phosphate uridylyltransferase |
| M. smegmatis MC2 155 | MSMEG_0384 | rfbA | 2e-09 | 22.92% (253) | glucose-1-phosphate thymidylyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1020 | galU | 1e-140 | 83.06% (301) | UTP--glucose-1-phosphate uridylyltransferase GalU |
| M. gilvum PYR-GCK | Mflv_1897 | - | 1e-152 | 90.85% (295) | nucleotidyl transferase |
| M. tuberculosis H37Rv | Rv0993 | galU | 1e-140 | 82.72% (301) | UTP--glucose-1-phosphate uridylyltransferase GalU |
| M. leprae Br4923 | MLBr_00182 | galU | 1e-144 | 85.57% (298) | putative UTP-glucose-1-phosphate uridylyltransferase |
| M. abscessus ATCC 19977 | MAB_1086 | - | 1e-145 | 87.07% (294) | putative UTP-glucose-1-phosphate uridylyltransferase (GalU) |
| M. marinum M | MMAR_4521 | galU | 1e-143 | 85.38% (301) | UTP--glucose-1-phosphate uridylyltransferase GalU |
| M. avium 104 | MAV_1102 | - | 1e-140 | 83.89% (298) | GalU protein |
| M. thermoresistible (build 8) | TH_0940 | galU | 1e-149 | 88.40% (293) | PROBABLE UTP--GLUCOSE-1-PHOSPHATE URIDYLYLTRANSFERASE GALU |
| M. ulcerans Agy99 | MUL_4693 | galU | 1e-144 | 85.71% (301) | UTP--glucose-1-phosphate uridylyltransferase GalU |
| M. vanbaalenii PYR-1 | Mvan_4833 | - | 1e-151 | 90.20% (296) | nucleotidyl transferase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4521|M.marinum_M ----MSQEVAIPFTAIVPAAGLGTRFLPATKTVPKELLPVVDTPGIELVA
MUL_4693|M.ulcerans_Agy99 ----MSQEVAIPFTAIVPAAGLGTRFLPATKTVPKELLPVVDTPGIELVA
Mb1020|M.bovis_AF2122/97 ---MSRPEVLTPFTAIVPAAGLGTRFLPATKTVPKELLPVVDTPGIELVA
Rv0993|M.tuberculosis_H37Rv ---MSRPEVLTPFTAIVPAAGLGTRFLPATKTVPKELLPVVDTPGIELVA
MLBr_00182|M.leprae_Br4923 MSLLLSAQVPIPFTAIVPAAGLGTRFLPATKTVPKELLPVVDTPGIELVA
MAV_1102|M.avium_104 --------MVVPRTAIVPAAGLGTRFLPATKTVPKELLPVVDTPGIELVA
MSMEG_5471|M.smegmatis_MC2_155 ---MS-----IPHTAVVPAAGLGTRFLPATKTVPKELLPVVDTPGIELVA
TH_0940|M.thermoresistible__bu ---MSESRAPVPRTAVVPAAGLGTRFLPVTKTVPKELLPVVDTPGIELVA
Mflv_1897|M.gilvum_PYR-GCK ---MNRPEVPVPRTAVVPAAGLGTRFLPATKTVPKELLPVVDTPGIELVA
Mvan_4833|M.vanbaalenii_PYR-1 ---MNRAEVPIPRTAVVPAAGLGTRFLPATKTVPKELLPVVDTPGIELVA
MAB_1086|M.abscessus_ATCC_1997 ---MTGPDVLIPRTAVVPAAGLGTRFLPATKTVPKELLPVVDTPGIELVA
* **:************.*********************
MMAR_4521|M.marinum_M AEAAAAGAERLVIVTSEGKDGVVAHFVEDLVLEGTLEARGKTAMLAKVRR
MUL_4693|M.ulcerans_Agy99 AEAAAAGAERLVIVTSEGKDGVVAHFVEDLVLEGTLEARGKTAMLAKVRR
Mb1020|M.bovis_AF2122/97 AEAAAAGAERLVIVTSEGKDGVVAHFVEDLVLEGTLEARGKIAMLAKVRR
Rv0993|M.tuberculosis_H37Rv AEAAAAGAERLVIVTSEGKDGVVAHFVEDLVLEGTLEARGKIAMLAKVRR
MLBr_00182|M.leprae_Br4923 AEAAAAGAERLVIVTSEGKDGVVAHFVQDLVLEGTLLARGNKAMLAKVRR
MAV_1102|M.avium_104 AEAAEAGAERLVIVTSEGKDGVVAHFVEDLVLESTLEERGKTAMLDKVRR
MSMEG_5471|M.smegmatis_MC2_155 AEAAEAGAERLVIVTSEGKDGVVAHFVQDLVLEGTLEARGKKTMLEKVRR
TH_0940|M.thermoresistible__bu EEAAEAGAQRLIIITSEGKDSVVAHFVQDLVLEGVLEARGKKTMLEKVRR
Mflv_1897|M.gilvum_PYR-GCK AEAAEAGAERLIIITSEGKDGVVAHFVEDLVLEGTLEARGKKTMLEKVRR
Mvan_4833|M.vanbaalenii_PYR-1 AEAAEAGAERLIIVTSEGKDGVVAHFVEDLVLEGTLEARGKKTMLEKVRR
MAB_1086|M.abscessus_ATCC_1997 EEAAEAGAERLVIVTSEGKDGVVAHFVEDLVLEGTLEARGKHAMLEKVRK
*** ***:**:*:******.******:*****..* **: :** ***:
MMAR_4521|M.marinum_M APALIKVESVVQAEPLGLGHAISCVEPTLSDDEDAVSVLLPDDLVLPTGV
MUL_4693|M.ulcerans_Agy99 APALIKVESVVQAEPLGLGHAISCVEPTLSDDEDAVSVLLPDDLVLPTGV
Mb1020|M.bovis_AF2122/97 APALIKVESVVQAEPLGLGHAIGCVEPTLSPDEDAVAVLLPDDLVLPTGV
Rv0993|M.tuberculosis_H37Rv APALIKVESVVQAEPLGLGHAIGCVEPTLSPDEDAVAVLLPDDLVLPTGV
MLBr_00182|M.leprae_Br4923 APELIKVESVVQAEPLGLGHAIGCAEPTLAPDEDAVSVLLPDDLVLPTGV
MAV_1102|M.avium_104 APRLIKVESVVQSEPLGLGHAIGCVEPRLADDEDAVAVLLPDDLVLPTGV
MSMEG_5471|M.smegmatis_MC2_155 APALIKVESVVQAEPLGLGHAVSCVESVLAADEDAISVLLPDDLVLPTGV
TH_0940|M.thermoresistible__bu APELIRVESVIQQEPLGLGHAVSCVESSLEPDEDAIAVLLPDDLVLPTGI
Mflv_1897|M.gilvum_PYR-GCK APALIKVESVVQAEPLGLGHAVGCVEASLAPDEDAIAVLLPDDLVLPTGV
Mvan_4833|M.vanbaalenii_PYR-1 APALIKVESVVQAEPLGLGHAISCVEASLAPDEDAIAVLLPDDLVLPTGV
MAB_1086|M.abscessus_ATCC_1997 APALIKVQSVVQAEPLGLGHAVGCVESVLDDDEDAVAVLLPDDLVWPRGV
** **:*:**:* ********:.*.*. * ****::******** * *:
MMAR_4521|M.marinum_M LETMSKVRARLGGTVLCAIEVSPDEISAYGVFDVEPVPGGEWAANPDVLK
MUL_4693|M.ulcerans_Agy99 LETMSKVRARLGGTVLCAIEVSPDEISAYGVFDVEPVPGGEWAANPNVLK
Mb1020|M.bovis_AF2122/97 LETMSKVRASRGGTVLCAIEVAREEISAYGVFDVEPVPDGDYTDDPNVLK
Rv0993|M.tuberculosis_H37Rv LETMSKVRASRGGTVLCAIEVAREEISAYGVFDVEPVPDGDYTDDPNVLK
MLBr_00182|M.leprae_Br4923 LETMSKVRASRGGTVLCAIEVASEEISSYGVFDVEPVPGG---DNPNVLK
MAV_1102|M.avium_104 LGTMSKVRAHYGGTVLCAIEVTPEEISAYGVFDVEPVPDD----DPDVLR
MSMEG_5471|M.smegmatis_MC2_155 LETMSKVRAKRGGSVLCAIEVPGDKISAYGVFDVETVPDAA---NPNVLR
TH_0940|M.thermoresistible__bu LETMSRVRAKRGGSVLCAIEVPGDEISAYGVFDVEPVPDST---NPDVLR
Mflv_1897|M.gilvum_PYR-GCK LETMAKVRAKRGGSVLCAIEVDREDISAYGVFDVETVPDAA---NPNVLR
Mvan_4833|M.vanbaalenii_PYR-1 LETMSKVRAKRGGSVLCAIEVDREDISAYGVIDVEPLPDAN---NPNVLK
MAB_1086|M.abscessus_ATCC_1997 LETMAKVRAKRGGSVLCAIEVPPHEISSYGAFDVEIVPDAA---NPNVLR
* **::*** **:******* ..**:**.:*** :*. :*:**:
MMAR_4521|M.marinum_M VKGMVEKPKTQDAPSMFAAAGRYVLDRAIFDALRRIERGAGGEVQLTDAI
MUL_4693|M.ulcerans_Agy99 VKGMVEKPKTQDAPSMFAAAGRYVLDRAIFDALRRIERGAGGEVQLTDAI
Mb1020|M.bovis_AF2122/97 VRGMVEKPKAETAPSRYAAAGRYVLDRAIFDALRRIDRGAGGEVQLTDAI
Rv0993|M.tuberculosis_H37Rv VRGMVEKPKAETAPSRYAAAGRYVLDRAIFDALRRIDQGAGGEVQLTDAI
MLBr_00182|M.leprae_Br4923 VIGMVEKPKAEDAPSTFAAAGRYVLDRAIFDALRRVSRGTGGEVQITDAI
MAV_1102|M.avium_104 VKGMVEKPKAEDAPSMFAAAGRYVLDRAIFDALRRIDRGAGGEVQLTDGI
MSMEG_5471|M.smegmatis_MC2_155 VKGMVEKPRPEDAPSHFAAAGRYVLDRAIFDALRRVPRGTGGEIQLTDAI
TH_0940|M.thermoresistible__bu VKGMVEKPKPEDAPSPYAAAGRYVLDRAIFDALRRVPRGAGGEIQLTDAI
Mflv_1897|M.gilvum_PYR-GCK VKGMVEKPKAEDAPSLFAAAGRYILDRAIFDALKRIPRGAGGELQLTDAI
Mvan_4833|M.vanbaalenii_PYR-1 VKGMVEKPKPEDAPSLYAAAGRYVLDRAIFDALRRIPRGAGGELQLTDAI
MAB_1086|M.abscessus_ATCC_1997 VKGMVEKPKTEEAPSNYAAAGRYLLDRAIFDALKRIPRGAGGELQLTDAI
* ******:.: *** :******:*********:*: :*:***:*:**.*
MMAR_4521|M.marinum_M ALLIAEGHPVHVVVHRGSRHDLGNPGGYLKAAVDFALDRDDYGPDLRRWL
MUL_4693|M.ulcerans_Agy99 ALLIAEGHPVHVVVHRGSRHDLGNPGGYLKAAVDFALDRDDYGPDLRRWL
Mb1020|M.bovis_AF2122/97 ALLIAEGHPVHVVVHQGSRHDLGNPGGYLKAAVDFALDRDDYGPDLRRWL
Rv0993|M.tuberculosis_H37Rv ALLIAEGHPVHVVVHQGSRHDLGNPGGYLKAAVDFALDRDDYGPDLRRWL
MLBr_00182|M.leprae_Br4923 ALLIKEGHPVHVVVHRGSRHDLGNPGGYLKAAVDFALDRDDYGPDLRRWL
MAV_1102|M.avium_104 ALLISEGHPVHVVVHRGSRHDLGNPGGYLKAAVDFALDRDDYGPDLRQWL
MSMEG_5471|M.smegmatis_MC2_155 ALLIEEDHPVHVVVHRGARHDLGNPGGYLKAAVDFALERDDYGPELRQWL
TH_0940|M.thermoresistible__bu ALLIEEGHPVHAVVHRGTRHDLGNPGGYLKAAVDFALERDDYGPDLREWL
Mflv_1897|M.gilvum_PYR-GCK ALLIEEGHPVHVVVHRGSRHDLGNPGGYLKAAVDFALERDDYGPELRRWL
Mvan_4833|M.vanbaalenii_PYR-1 ALLIQEGHPVHVVIHRGSRHDLGNPGGYLKAAVDFALERDDYGPELRRWL
MAB_1086|M.abscessus_ATCC_1997 ALLIDEGHPVHVVVHRGSRHDLGNPGGYLKAAVDFALDRDDYGPGLREWL
**** *.****.*:*:*:*******************:****** **.**
MMAR_4521|M.marinum_M VARLGLTEQ------
MUL_4693|M.ulcerans_Agy99 VARLGLTEQ------
Mb1020|M.bovis_AF2122/97 VARLGLTEQ------
Rv0993|M.tuberculosis_H37Rv VARLGLTEQ------
MLBr_00182|M.leprae_Br4923 VERLGLIEQ------
MAV_1102|M.avium_104 VARLGLSEQ------
MSMEG_5471|M.smegmatis_MC2_155 VERLGLAEK------
TH_0940|M.thermoresistible__bu LERLGLT--------
Mflv_1897|M.gilvum_PYR-GCK VERLGLPAGTEG---
Mvan_4833|M.vanbaalenii_PYR-1 VERLGLAPATSGQ--
MAB_1086|M.abscessus_ATCC_1997 VERLGRESSVLPESE
: ***