For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PEVVFSVDHSKSMRDQAVPGHNRWHPDIPAAATVKPGSEFRIECKEWTDGQIGNNDSANDVRDVDLAPCH MLSGPIKVEGAEPGDLLIVDILDIGPVPQTNGPNCGEGWGYSGIFAKVNGGGFLTDYYPDAYKAIWDFHG QQCTSRHVPGVRYTGITHPGLFGTAPSPDLLAKWNERERALIATDPDRVPPLALPPLVDGTLGGTASGDL LQAIANDGARTVPPRENGGNHDIKNFTRGSRIFYPVFVEGAMLSGGDLHFSQGDGEINFCGAIEMGGFID MHVDLIKGGMETYGVTTNPIFMPGRVEPLYSEWLTFIGISVDHAENRNAYMDATMAYRNACLNAIEYLKK WGYTGEQAYLILGTSPIEGRIGGVVDIPNACCSVFLPTEIFDFDIRPGGKGPQKVDRGQVAVTS*
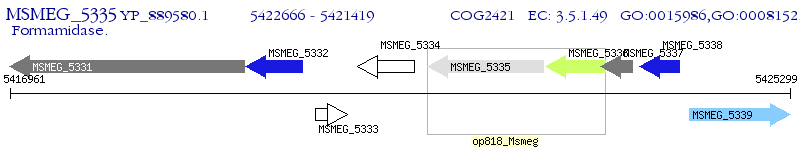
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5335 | - | - | 100% (415) | formamidase |
| M. smegmatis MC2 155 | MSMEG_4367 | - | e-162 | 64.41% (413) | formamidase |
| M. smegmatis MC2 155 | MSMEG_0484 | - | 3e-49 | 34.25% (400) | formamidase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2015 | - | 1e-11 | 28.97% (214) | acetamidase/formamidase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0587 | - | 1e-165 | 64.66% (416) | acetamidase/formamidase |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_0852 | - | 1e-11 | 29.03% (217) | acetamidase/Formamidase family protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0290 | - | 3e-10 | 39.53% (86) | acetamidase/formamidase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2015|M.gilvum_PYR-GCK MTDPARRAAETLWTGIGRRDFLRAVAAVSAGAGVAGVSSACASGTPGPAS
TH_0852|M.thermoresistible__bu ---------------------------LSAGAGIATTLGACSS----RST
Mvan_0290|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5335|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0587|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_2015|M.gilvum_PYR-GCK QSPSSAATDVAILQPGEGDPVGDHYLQSAPDEVLWGYVPNVHAAPVMRMS
TH_0852|M.thermoresistible__bu VRATGASTDFEILQPGQGEITGDHYLQSVPDQVLWGYVPTVHATPVMQMR
Mvan_0290|M.vanbaalenii_PYR-1 -------MIFPTLHPGEGHIGGEHYLPSHPDEVLWGWLPNAQTRPALSVA
MSMEG_5335|M.smegmatis_MC2_155 MPEVVFSVDHSKSMRDQAVPGHNRWHPDIPAAATVK--PGSEFRIECKEW
MAB_0587|M.abscessus_ATCC_1997 MPEVLFPLDSNKKFTEQQIIGHNRWHPDIPPAAVVK--PGTSFRVHCREW
: ::: . * . *
Mflv_2015|M.gilvum_PYR-GCK SGQTVTIDAVSHEGILEDQGRDPVGYFGGQGVSESEVLRDAIAIAAEYDR
TH_0852|M.thermoresistible__bu SGQTVTIDTVSHEGILEDQGRNPVEYFGAEGVDENDVLEDVIAVAAEYDR
Mvan_0290|M.vanbaalenii_PYR-1 SGDTVTIDTVSHEGILEDQGRDPVSYFAQFGVTN--VLKDVIDIASS---
MSMEG_5335|M.smegmatis_MC2_155 TDGQIGNNDSAND--VRDVDLAPCHMLSG-PIKVEGAEPGDLLIVDILDI
MAB_0587|M.abscessus_ATCC_1997 FDGAIHNDDSADD--IRDAPLTTVHALSG-PFSVEGAQPGDLLIVDILDV
. : : :.: :.* . :. . . . : :.
Mflv_2015|M.gilvum_PYR-GCK TPRNFDKDGPHVVTGPVFVEGAQPGDVLKIEILQAIPRVPYGVVSSRHGK
TH_0852|M.thermoresistible__bu TPRDFDKDGPHVVTGPIFIEGAEPGDVLKIETLEATPRVPYGVVSSRHGK
Mvan_0290|M.vanbaalenii_PYR-1 PLAHDDSCGPHIVTGPVRVANADPGDVLKIEVLDLAMRVPYGFVSSRHGY
MSMEG_5335|M.smegmatis_MC2_155 GPVPQTN-GPNCGEG--WGYSGIFAKVNGGGFLTDYYPDAYKAIWDFHGQ
MAB_0587|M.abscessus_ATCC_1997 GPIPQEDSGPLAGQG--WGYTGIFPTVNGGGFLTEQFPDAYKAIWDFSGQ
. ** * . * * .* : . *
Mflv_2015|M.gilvum_PYR-GCK GALARTADGAAPAGVGLSEVMPPVATDGRPDPDPTRYGNVSTFTAVEDGH
TH_0852|M.thermoresistible__bu GALALTADGTPPAGITLDEVMPPAVTDGRSNPDPTRYGNLSTFTAVEDGH
Mvan_0290|M.vanbaalenii_PYR-1 GALAGEFP-EFPDSEPVREVDQIVSMGTICHFSWVEMLQGKPYGKIAAGH
MSMEG_5335|M.smegmatis_MC2_155 QCTSRHVP-----GVRYTGITHPGLFGTAPSPDLLAKWNERERALIATDP
MAB_0587|M.abscessus_ATCC_1997 TATSRHVP-----GVRFTGIVHPGLMGTAPSAELLARWNAREGALIATDP
. : . : . . : : .
Mflv_2015|M.gilvum_PYR-GCK GVMRYGSAQVRFPLRPFMGMMGVAFTQDREITAATAN-SIPPTVGGGNID
TH_0852|M.thermoresistible__bu GVMSYGDARVRFPLAPFMGIMGVAFAQDRDPTAPTAN-SVPPTIAGGNID
Mvan_0290|M.vanbaalenii_PYR-1 GSGR----TVQFPVNPFLGIMGVARA-----TEDPVP-SVPPGTHGGNID
MSMEG_5335|M.smegmatis_MC2_155 DRVP----PLALPPLVDGTLGGTASGDLLQAIANDGARTVPPRENGGNHD
MAB_0587|M.abscessus_ATCC_1997 NRVP----ALALPPEPRDAVLGALSGDSFDRAAAEAARTAPPRENGGNQD
. : :* : *. : ** *** *
Mflv_2015|M.gilvum_PYR-GCK IRLLGEGATFFLPVFAEGALFYVGDPHMAMGDGEVALT-AMEGSLRGTYR
TH_0852|M.thermoresistible__bu IRLLGEGSTLYLPVFAEGALFYVGDPHMAMGDGEVALT-AMEGSLRGTYR
Mvan_0290|M.vanbaalenii_PYR-1 IKHVVTGSTLYLPVQVEGANFFAGDPHFAQGNGEVALT-ALEASLRATVR
MSMEG_5335|M.smegmatis_MC2_155 IKNFTRGSRIFYPVFVEGAMLSGGDLHFSQGDGEINFCGAIEMGGFIDMH
MAB_0587|M.abscessus_ATCC_1997 IKNLTKGSRVFYPVFVPGGKLSVGDLHFSQGDGEITFCGAIEMGGFIDLH
*: . *: .: ** . *. : ** *:: *:**: : *:* . :
Mflv_2015|M.gilvum_PYR-GCK LTVCKPGSGDAPSVAYRYPFGETVD----AWVP-IGLSDPDGSVGGQGSD
TH_0852|M.thermoresistible__bu LTVCKPGSGDAPSVAYHYPFAETGD----AWVP-IGLSDPDGSVGGQGTD
Mvan_0290|M.vanbaalenii_PYR-1 LTVLKSGDAHSAIGALTNPVVETAT----HWIP-TGMD----------AD
MSMEG_5335|M.smegmatis_MC2_155 VDLIKGGMETYGVTTNPIFMPGRVEPLYSEWLTFIGISVDHAENRNAYMD
MAB_0587|M.abscessus_ATCC_1997 VDVIKGGMDTYGVFENAIFMPGNTDPQYSQWLAFSGTSVTLDDEQ-RYLD
: : * * . *:. * . *
Mflv_2015|M.gilvum_PYR-GCK LDVAMRRAVVNALDFLETDQGMDRATAYAYLSAAADFSVSQVVDRTVGVH
TH_0852|M.thermoresistible__bu LDVAMRRAVANALDFLENDRGMDRATAYAYLSAASHFAISQVVDRTVGVH
Mvan_0290|M.vanbaalenii_PYR-1 LDEAMRIAVRNAIAFLNTRFGVPRDVALAYLSAAGDFEVSQVVDAVKGVH
MSMEG_5335|M.smegmatis_MC2_155 ATMAYRNACLNAIEYLKKWGYTGEQAYLILGTSPIEGRIGGVVDIPNACC
MAB_0587|M.abscessus_ATCC_1997 SHLAYQRACLHAIDYLTKFGYSPEQAYLLLGAAPIEGRLSGVVDIPNSCA
* : * :*: :* . . . ::. . :. *** .
Mflv_2015|M.gilvum_PYR-GCK GQIYKGHFGQS------------------------
TH_0852|M.thermoresistible__bu GQIFKSHFAA-------------------------
Mvan_0290|M.vanbaalenii_PYR-1 CMIRKADWAAWL-----------------------
MSMEG_5335|M.smegmatis_MC2_155 SVFLPTEIFDFDIRPGGKGPQKVDRGQVAVTS---
MAB_0587|M.abscessus_ATCC_1997 TVYIPTAIFDFPVTPTASGPVRIDPGPGAPRSVAR