For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MCEHLRMDPQDDPEARIRDLERSLAERAKELGTGADEGHAPPAPPPPPPTQPWTQTFGTAYPPPPPGPPA PWPGYAEDFPAPPRQHVPVAHTGRILMILVAVAVFLITTGIAAFLMFTNTVTSSLQPVADNPAGRPALPY SPPSNPVFPAIETPSVAGPGEVINVAGASNDREVTCDGGTVMVSGVENTITITGHCAAVTISGIRNVIEV DEADTIGVSGFENRVTYSSGEPEITKSGMDNTVERRAP
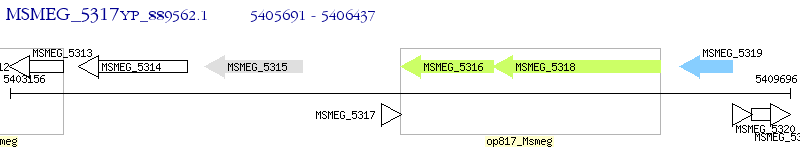
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5317 | - | - | 100% (248) | hypothetical protein MSMEG_5317 |
| M. smegmatis MC2 155 | MSMEG_1393 | - | 2e-10 | 35.23% (88) | hypothetical protein MSMEG_1393 |
| M. smegmatis MC2 155 | MSMEG_1394 | - | 9e-09 | 28.57% (105) | transmembrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0322c | - | 9e-36 | 37.55% (245) | hypothetical protein Mb0322c |
| M. gilvum PYR-GCK | Mflv_5128 | - | 2e-42 | 38.40% (250) | hypothetical protein Mflv_5128 |
| M. tuberculosis H37Rv | Rv0314c | - | 3e-36 | 37.96% (245) | hypothetical protein Rv0314c |
| M. leprae Br4923 | MLBr_02395 | Ag36 | 4e-06 | 33.33% (108) | proline rich antigenic protein |
| M. abscessus ATCC 19977 | MAB_1176c | - | 1e-29 | 34.41% (247) | hypothetical protein MAB_1176c |
| M. marinum M | MMAR_0565 | - | 6e-36 | 38.06% (247) | hypothetical protein MMAR_0565 |
| M. avium 104 | MAV_4846 | - | 3e-36 | 36.36% (242) | hypothetical protein MAV_4846 |
| M. thermoresistible (build 8) | TH_1896 | - | 4e-11 | 36.47% (85) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_4742 | - | 2e-38 | 39.29% (252) | hypothetical protein MUL_4742 |
| M. vanbaalenii PYR-1 | Mvan_1215 | - | 2e-41 | 39.76% (249) | hypothetical protein Mvan_1215 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0322c|M.bovis_AF2122/97 MIVVWEHLCMNPEDDPEARIRELERPLADVARASELGGSQSG--------
Rv0314c|M.tuberculosis_H37Rv MIVVWEHLCMNPEDDPEARIRELERPLADVARASELGGSQSG--------
MMAR_0565|M.marinum_M ---------MNPEDDPEARIRQLEQPLADAARASELGSSQPAGESAYQNY
MUL_4742|M.ulcerans_Agy99 ---MWEHLCMNPEDDPEARIRQLEQPLADAARASELGSSQPAGESAYQNY
MAV_4846|M.avium_104 ---------MAANDDPEERIRELERPLADSARASEQGSAPPTG-------
MAB_1176c|M.abscessus_ATCC_199 ---------MEPDGDPEARIRELERPLSERAQRSELGAPSQS--------
Mflv_5128|M.gilvum_PYR-GCK ---MCQHLRMEPNGDPEARIRDLERPLADRARASELGTHPYGAPPVPPYQ
Mvan_1215|M.vanbaalenii_PYR-1 ---MCKHLPMEPSGDPEARIRDLERPLADQAAANELGTHAY--------Q
TH_1896|M.thermoresistible__bu --------------------------------------------------
MSMEG_5317|M.smegmatis_MC2_155 ---MCEHLRMDPQDDPEARIRDLERSLAERAKELGTGADEGHAPPAPPPP
MLBr_02395|M.leprae_Br4923 --MTDQPPPSGSNPTPAPPPPGSSGGYEPSFAPSELGSAYPPPTAPPVGG
Mb0322c|M.bovis_AF2122/97 ----------GYTYPPGPPPPP---YSYGGPFGGPSPRSSSGNRAWWILA
Rv0314c|M.tuberculosis_H37Rv ----------GYTYPPGPPPPP---YSYGGPFGGPSPRSSSGNRAWWILA
MMAR_0565|M.marinum_M QGYQGYQGYPGYQPPPGSVPPS---YGYQPAFPGATPRSSSGNRAFWIIA
MUL_4742|M.ulcerans_Agy99 QG------YPGYQPPPGSVPPSPPPYGYQPAFPGATPRSSSGNRAFWIIA
MAV_4846|M.avium_104 ---------PGYPPAPAP-------WSYGGPVPGPPGQRPSGNRVWWVVG
MAB_1176c|M.abscessus_ATCC_199 -------------PYPLP--------AYDTTQISRSPVRTFR-RRFLVFL
Mflv_5128|M.gilvum_PYR-GCK DVALP--PMPPGVEHPAPVYGS-----PYYAPPQRVVHKRSN-TALWLIP
Mvan_1215|M.vanbaalenii_PYR-1 DVAPPTTPVPPYPYPPYAQYGS-----PYYAPPQRVVDKRSHTTALWLIP
TH_1896|M.thermoresistible__bu -------------------------------------------MGHDVIR
MSMEG_5317|M.smegmatis_MC2_155 PPTQPWTQTFGTAYPPPPPGPPAPWPGYAEDFPAPPRQHVPVAHTGRILM
MLBr_02395|M.leprae_Br4923 SYPPPPPPGGSYPPPPPPGGSYPPPPPSTGAYAPPPPGPAIRSLPKEAYT
Mb0322c|M.bovis_AF2122/97 AVVVVGVLVLVGGIAAFSAQRLSQGNFVVLSPTPSVSRAVP---------
Rv0314c|M.tuberculosis_H37Rv AVVVVGVLVLVGGIAAFSAQRLSQGNFVVLSPTPSVSRAVP---------
MMAR_0565|M.marinum_M AVFVVGILGAVAAVALFAVNRVSEP-FTTLSPGTTVISTPPGPMSPGNKT
MUL_4742|M.ulcerans_Agy99 AVFVVGILGAVAAVALFAVNRVSEP-FTTLSPGTTVISTPPGPMSPGNKT
MAV_4846|M.avium_104 TLIAVGVLALAGGIAAFAAHQLSGVKSIINSPPTISRTFGPPPGISTTPA
MAB_1176c|M.abscessus_ATCC_199 AVITAG----AAGLVFYAAEPTIRQGGPTVSGPSGTRPARSTVVKIPRAT
Mflv_5128|M.gilvum_PYR-GCK VVVVVVLAAGIIGIVAYFSSGTPDSYT--YTPPVSGGGGPLDAP-LPPQV
Mvan_1215|M.vanbaalenii_PYR-1 LVVVGAIVAGAVAVVVFFNTGSPDSATPIPVPPISGGGGPLDP---PPSV
TH_1896|M.thermoresistible__bu TVIGAFAAAAALGLAG---CGSQDGDT--PGPSATAG-------------
MSMEG_5317|M.smegmatis_MC2_155 ILVAVAVFLITTGIAAFLMFTNTVTSSLQPVADNPAGRPALPYS---PPS
MLBr_02395|M.leprae_Br4923 FWVTRVLAYVIDNIPATVLLGIGMLIQTLTKQEACVTDITQYNVNQYCAT
. :
Mb0322c|M.bovis_AF2122/97 -TPTAQPATTL-PPAGASLSVSGVNVNRTIACNDS-IVSVSGMSNTVVIT
Rv0314c|M.tuberculosis_H37Rv -TPTAQPATTL-PPAGASLSVSGVNVNRTIACNDS-IVSVSGMSNTVVIT
MMAR_0565|M.marinum_M QTPTAGPSTSAGPPAGSTISVVGVNQSQTITCNDN-IVNVSGMSNTVTIT
MUL_4742|M.ulcerans_Agy99 QTPTAGPSTSAGPPAGSTISVVGVNQSQTITCNDN-IVNVSGMSNTVTIT
MAV_4846|M.avium_104 PSTSRRTPTSPAAPPGTELSVAGINENRTLACDDN-PVSVSGVSNTVVIT
MAB_1176c|M.abscessus_ATCC_199 PSSTAAPGPTF-VPAGSTFSVAGTHKNVAISCDGC-SVSVSGVSHTVEIL
Mflv_5128|M.gilvum_PYR-GCK QTRTDSTEQVITVDSGGFVSIGGVEKNQTIVCDEG-TVSISGVNNTIEVT
Mvan_1215|M.vanbaalenii_PYR-1 QTRIESTEQIVTVESGGFVSLGGVETRQTVVCDDG-TVNISGVNNTIEVQ
TH_1896|M.thermoresistible__bu -------EDGVRVEIANTINYGSFGTTTEIDCADGKSLNIGGSNNTLTVL
MSMEG_5317|M.smegmatis_MC2_155 NPVFPAIETPSVAGPGEVINVAGASNDREVTCDGG-TVMVSGVENTITIT
MLBr_02395|M.leprae_Br4923 QPTGIGMLAFWFAWLMATAYLVWNYGYRQGATGSSIGKTVMKFKVISEAT
: .
Mb0322c|M.bovis_AF2122/97 GHCTSLTVSGMRNSVTADSVD-TIEAAGFNNEVTYHSGSPKISNAGGSNS
Rv0314c|M.tuberculosis_H37Rv GHCTSLTVSGMRNSVTVDSVD-TIEAAGFNNEVTYHSGSPKISNAGGSNS
MMAR_0565|M.marinum_M GHCASVSVSGLQNTVTVETAD-RITASGLNNKVTYQSGSPKISKSGSGNV
MUL_4742|M.ulcerans_Agy99 GHCASVSVSGLQNTVTVETAD-RITASGLNNKVTYQSGSPKISKSGSGNV
MAV_4846|M.avium_104 GHCTSLSVSGVQNTVTVEAVE-RIGASGFNNKVTYHSGTPSISNSGGSNV
MAB_1176c|M.abscessus_ATCC_199 GNCDTLTVSGVENTVTVETAV-KIGVSGIDNQVTYRTGEPQVAKSGNNNT
Mflv_5128|M.gilvum_PYR-GCK GRCGSVTVSGVDNTVTVESAG-TISASGFHNRVTFLAGDPQTSTSGSGNV
Mvan_1215|M.vanbaalenii_PYR-1 GACTMVTVSGVENTVTVESAD-TISASGFDNRVTYRTGTPEISKSGRGNV
TH_1896|M.thermoresistible__bu GTCANVNVGGTDNRITVERVDNEISVTGFNNTLTYRDGDPTVSDTGRGNN
MSMEG_5317|M.smegmatis_MC2_155 GHCAAVTISGIRNVIEVDEAD-TIGVSGFENRVTYSSGEPEITKSGMDNT
MLBr_02395|M.leprae_Br4923 GQPIGFGMSVVRQLAHFVDAV--ICCIGFLFPLWDSKRQTLADKIMTTVC
* . :. : . * *: : .
Mb0322c|M.bovis_AF2122/97 VQQG--
Rv0314c|M.tuberculosis_H37Rv VQQG--
MMAR_0565|M.marinum_M IEQG--
MUL_4742|M.ulcerans_Agy99 IEQG--
MAV_4846|M.avium_104 VGRG--
MAB_1176c|M.abscessus_ATCC_199 VEQS--
Mflv_5128|M.gilvum_PYR-GCK IERQ--
Mvan_1215|M.vanbaalenii_PYR-1 VEPG--
TH_1896|M.thermoresistible__bu IGRG--
MSMEG_5317|M.smegmatis_MC2_155 VERRAP
MLBr_02395|M.leprae_Br4923 LPI---
: