For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LKETPPAGWPQSLSDFSMTWTAEPGIDLTKRPAVVARAYIESFYLADFTDDERYLYPGFEGSVSRNQSDG PEGSDSLWPASDGADAVVGTFRQHLLRIDQSGQDVSVVGCMYTYDSALEVGGKFKANTENGPFGGINAFR VELQAPKDDDLLPPQQGPSRAPFGNVFGNWKVTNHQGGYLVTAEWPNHSEDNEQCLARSGVTPESRKFYP GTTYQRSDFPVLPATPGWPAESDTSTHEPR
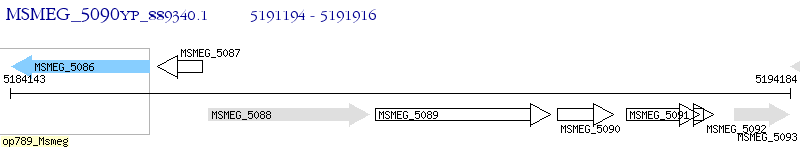
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5090 | - | - | 100% (240) | hypothetical protein MSMEG_5090 |
| M. smegmatis MC2 155 | MSMEG_5099 | - | 2e-12 | 27.24% (257) | hypothetical protein MSMEG_5099 |
| M. smegmatis MC2 155 | MSMEG_5097 | - | 1e-06 | 26.11% (180) | hypothetical protein MSMEG_5097 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1284c | - | 7e-22 | 29.32% (266) | hypothetical protein MAB_1284c |
| M. marinum M | MMAR_3060 | - | 3e-55 | 46.22% (238) | hypothetical protein MMAR_3060 |
| M. avium 104 | MAV_4813 | - | 7e-43 | 42.11% (209) | hypothetical protein MAV_4813 |
| M. thermoresistible (build 8) | TH_3379 | - | 4e-06 | 32.05% (78) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_2304 | - | 3e-55 | 47.70% (239) | hypothetical protein MUL_2304 |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3060|M.marinum_M -MRIRSELRSLAAFLAVAVLLVGGCGSGGRSGEGPASTSAAPPAGWPAAL
MUL_2304|M.ulcerans_Agy99 MMRIRPELRTIAAFLAVAVLLVGGCGSGGRSGEGPASTSAVAPAGWPAAL
MAV_4813|M.avium_104 --------------------------------------------------
MSMEG_5090|M.smegmatis_MC2_155 -------------------------------------MKETPPAGWPQSL
MAB_1284c|M.abscessus_ATCC_199 --------MTHPRKRHAGARILFGLLATIVSMSAGCTPTEQPPIPFTGLT
TH_3379|M.thermoresistible__bu --------------------------------------------------
MMAR_3060|M.marinum_M GDFTMVWTAEPGVDVTAWPAVVVRAYTESFVLASIMNDDKYLYPGFRQSV
MUL_2304|M.ulcerans_Agy99 DDFTMVWTAEPGIDVTAWPAVVVRAYTESFLLASITRDDKYLYPGFAQSV
MAV_4813|M.avium_104 --------------------MPVRAYTESYLLASIMGDNRYLYPGFQQSV
MSMEG_5090|M.smegmatis_MC2_155 SDFSMTWTAEPGIDLTKRPAVVARAYIESFYLADFTDDERYLYPGFEGSV
MAB_1284c|M.abscessus_ATCC_199 KDLRIRWSAEPGIDLLTGPAVIVRAYRESYVVGGLMANPAYYYPGFEHAV
TH_3379|M.thermoresistible__bu ----------------------------------------MPYLGAMVGD
* * .
MMAR_3060|M.marinum_M DPNKSIDDPIGT-----QYLWPRKYYPQRP-WVGTQREHILSVT-ASGRD
MUL_2304|M.ulcerans_Agy99 DPNKSIHDPIST-----QFLWPKTDEPREPRWVGTQRDHILSVT-TSGRD
MAV_4813|M.avium_104 DPNQSINHPIGT-----QSLWPKTNAPQQQ-WIGTVQVHILSVT-TAGRD
MSMEG_5090|M.smegmatis_MC2_155 SRNQS-DGPEGS-----DSLWPASDGADAV--VGTFRQHLLRID-QSGQD
MAB_1284c|M.abscessus_ATCC_199 PQNGPNVSPLIRPLVTGDSQLERRGFQIATPIVGTWREHILSLTGDPTSG
TH_3379|M.thermoresistible__bu DRNTAGFTPLDG--------------------------------------
* . *
MMAR_3060|M.marinum_M ITAIVCEYTFGAASEGRF-GRRYAPHAVGPDPYKG-----IESLRITMTA
MUL_2304|M.ulcerans_Agy99 VTVIVCEYTFGVASESYLRGGWWRAHVVGPEPYSG-----IDKMRITMTA
MAV_4813|M.avium_104 VTVVACEYTFGSAQPARH---GYEPNIGKPPPYSG-----IDPLRITMET
MSMEG_5090|M.smegmatis_MC2_155 VSVVGCMYTYDSALEVGG---KFKANTEN-GPFGG-----INAFRVELQA
MAB_1284c|M.abscessus_ATCC_199 YTAKVCVWDTGTGVLAPNGQYRYPNRLPSNSMWSVRAGVGTLAVRITITP
TH_3379|M.thermoresistible__bu ---------------------------------------------LHSEM
:
MMAR_3060|M.marinum_M P--AKPVLPLPAQQGPARAPSVDVFAGWRITSHQ-GGWFARSGVGSDWPH
MUL_2304|M.ulcerans_Agy99 P--VKPVAPLPAQQGPARAPSIDVFAGWRITSRQ-GGWFVAAGVGSDWPG
MAV_4813|M.avium_104 P--AKP-GPQSPQRGPARSPSVDVFNGWRITSHQ-GGYFAQSGVGDEWPN
MSMEG_5090|M.smegmatis_MC2_155 P--KDD-DLLPPQQGPSRAPFGNVFGNWKVTNHQ-GGYLVTA----EWPN
MAB_1284c|M.abscessus_ATCC_199 PPPFEPDPTKKSQQGSAPNPSTDIFGGWKIRAADSLGIASWMDQRSDWPL
TH_3379|M.thermoresistible__bu P-----------RSAALFAPAIDVSGDWTITGYL---NWFWVYQTPEWPH
* : .. * :: .* : :**
MMAR_3060|M.marinum_M VIE--DQDTCLAKAPPHPD----FVSGEEYDRSLFPTLPASPGWPSMSAN
MUL_2304|M.ulcerans_Agy99 AAE--DRDTCLAKAPPHPD----LVPGEEYDRSLFPTLPASPGWPSTSAN
MAV_4813|M.avium_104 AIE--DRNTCLAKASQHPD----LQRGGEYPRTDFPTLPPSPGWPAPSAA
MSMEG_5090|M.smegmatis_MC2_155 HSE--DNEQCLARSGVTPESR-KFYPGTTYQRSDFPVLPATPGWPAESDT
MAB_1284c|M.abscessus_ATCC_199 DDFRLTLNACGERAPDLLTGQGNFITVGDHPRSDFPTLPADPGWPAAGT-
TH_3379|M.thermoresistible__bu GQE--DLNARIDRAPDSHQHR-QLLIVGPHPRDYFPTEPAWPGWPRAVK-
: :: : : * **. *. ****
MMAR_3060|M.marinum_M A-----
MUL_2304|M.ulcerans_Agy99 A-----
MAV_4813|M.avium_104 S-----
MSMEG_5090|M.smegmatis_MC2_155 STHEPR
MAB_1284c|M.abscessus_ATCC_199 ------
TH_3379|M.thermoresistible__bu ------