For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVARVCWGAFLAVCAITLAGCSSDPVIAGRATSMLFVPDRVGGLQVTEGPSGTRPDAPAPRRQAENTDGG QIDNLALLAVDDIEDFWSQNYSGGSLRGSFTPIRRLVSYNSDQSPGIEICGEDTEGLTNAFYCFNDDVMA WDRGVAIPVAAQYFGQMGVVGVMAHEYGHAVQHQAGLIDTRSTPVLIAEQQADCFAGVYLRWVAAGNSPR FALSTGDGLNHVLAGIIYIRDPLMTQLDAALTGNEHGSALDRVSAFQIGFSGNVDQCAAIDMDDIKKRRG DLPKFLDGVVADPSAGNDEITEDLLTDVMDSLGQIFAPADPPSLTIGDASGCPDAQPTPPASYCPATNTI VVDMPGLQALGETKTENEEQELLQGDNSAISVLTSRYALAVAREKGHAIDNPVAAMRTACLTGVAQAAMA EPGLPVRLSAGDTDEAISGLLTNGLAASDVNGALLPAGFTRILGYRSGLQGDDAQCYQRFP
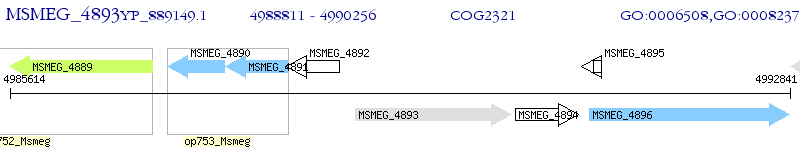
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4893 | - | - | 100% (481) | putative neutral zinc metallopeptidase |
| M. smegmatis MC2 155 | MSMEG_4913 | - | e-133 | 50.61% (488) | LpqM protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0427 | lpqM | 1e-133 | 53.18% (472) | lipoprotein peptidase LpqM |
| M. gilvum PYR-GCK | Mflv_2346 | - | 1e-127 | 49.06% (481) | LpqM |
| M. tuberculosis H37Rv | Rv0419 | lpqM | 1e-133 | 53.18% (472) | lipoprotein peptidase LpqM |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1470c | - | 1e-104 | 44.51% (483) | lipoprotein peptidase LpqM |
| M. marinum M | MMAR_0728 | lpqM | 1e-134 | 51.66% (482) | lipoprotein peptidase LpqM |
| M. avium 104 | MAV_4736 | - | 1e-135 | 53.99% (476) | LpqM protein |
| M. thermoresistible (build 8) | TH_1205 | - | 1e-125 | 50.21% (480) | LpqM protein |
| M. ulcerans Agy99 | MUL_3589 | lpqM | 1e-132 | 51.44% (486) | lipoprotein peptidase LpqM |
| M. vanbaalenii PYR-1 | Mvan_4299 | - | 1e-126 | 48.76% (482) | LpqM |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2346|M.gilvum_PYR-GCK --------MSPSRSSKPSRFRSSAALLAG-TAVLAGAACST--MLQGNAV
Mvan_4299|M.vanbaalenii_PYR-1 --------MSPSRRPG-TRIGVAAAVCA----VLAATACST--TLQGNAV
TH_1205|M.thermoresistible__bu ----------MTRRRRLAAFSTTPALLAVLVVVLLVSGCAR--VVRGSAV
Mb0427|M.bovis_AF2122/97 MHGRGRYRPLVRCVRPRRVAASVRTPIACLAAVVVIAGCTT--VVDGRAL
Rv0419|M.tuberculosis_H37Rv MHGRGRYRPLVRCVRPRRVAASVRTPIACLAAVVVIAGCTT--VVDGRAL
MMAR_0728|M.marinum_M MDNRGRKWLGVRRSRHGGLIASLWAATAFLTALLVVAGCTT--IVDGRAL
MUL_3589|M.ulcerans_Agy99 MDNRGRKWLGVRRSRHSGLIASLWAATAFLTALLVVAGCTT--IADGRAL
MAV_4736|M.avium_104 ---------------------------MSAAAVLLVAGCST--FVEGRAL
MSMEG_4893|M.smegmatis_MC2_155 ------------------MVARVCWGAFLAVCAITLAGCSSDPVIAGRAT
MAB_1470c|M.abscessus_ATCC_199 --------------MSIPLRWFHTSTTILACCVLGLAGCAN--TVDGTPV
: :.*: * .
Mflv_2346|M.gilvum_PYR-GCK SVFDDPFKVAGMPATDGPTGLRPDAESPARDVTGTDGGDIDNLAASAVSD
Mvan_4299|M.vanbaalenii_PYR-1 SVFDDPFRVAGMPATDGPTGVRPNAEPPARDVEGTDGGAIDELAASAISD
TH_1205|M.thermoresistible__bu SVFADPFRVAGMPAVDGPSGLRPDADAPTRTVRATDGGPIDQLAASAVSD
Mb0427|M.bovis_AF2122/97 SILNDPFRVGGLPATNGPSGARPDAPAASGTVINTNNGAIDKLSLLSVND
Rv0419|M.tuberculosis_H37Rv SILNDPFRVGGLPATNGPSGARPDAPAASGTVINTNNGAIDKLSLLSVND
MMAR_0728|M.marinum_M SILNDPFRVGGLPATNGPSGPRPDGPPPTGTVINTNNGPIDKLSLLSVND
MUL_3589|M.ulcerans_Agy99 SILNDPFRVGGLPATNGPSGPRPDGPPPTGTVINTNNGPIDKLSLLSVND
MAV_4736|M.avium_104 SMLNDPFLVGGLPATNGPSGIRSNAPAPTGKVLNTDNGSIDSLSLLSIND
MSMEG_4893|M.smegmatis_MC2_155 SMLFVPDRVGGLQVTEGPSGTRPDAPAPRRQAENTDGGQIDNLALLAVDD
MAB_1470c|M.abscessus_ATCC_199 SQLGNAYQIAGLPVVDGPSGLRPHPTSADRAVRGTDSGQMDRLAVNAIAD
* : . :.*: ..:**:* *.. .. . *:.* :* *: :: *
Mflv_2346|M.gilvum_PYR-GCK IEDYWATAYE-DAFDERFKPVAELISWDANGFDG-EFCGDTTYGLVNAGY
Mvan_4299|M.vanbaalenii_PYR-1 IEDYWAVAYE-ETFDEKFRPVAELISWDANGFDG-EFCGEDTYGLINAGY
TH_1205|M.thermoresistible__bu VEDYWDTAYL-DTFGERFTPVRGLVSWDAAELFE-VFCSMDTHGLENAAF
Mb0427|M.bovis_AF2122/97 IEDYWMAVYS-ESLKGTFRPVGKLVSYDSNDPSSPIVCHIDTYQLVNAFF
Rv0419|M.tuberculosis_H37Rv IEDYWMAVYS-ESLKGTFRPVGKLVSYDSNDPSSPIVCHIDTYQLVNAFF
MMAR_0728|M.marinum_M IQDYWKANY--EPLQGTFRPVSKLVSYDSNDPNSPIVCHNDTYELVNAFF
MUL_3589|M.ulcerans_Agy99 IQDYWKANY--EPLQGTFRPVSKLVSYDSNDPNSPIVCHNDTYELVNAFF
MAV_4736|M.avium_104 IEDYWRSVYG-QSLKGKFVPISKIVSYNSTDPSSPIVCHNDTYQLVNAFY
MSMEG_4893|M.smegmatis_MC2_155 IEDFWSQNYSGGSLRGSFTPIRRLVSYNSDQSPGIEICGEDTEGLTNAFY
MAB_1470c|M.abscessus_ATCC_199 IEDFWQEAYP-KTFWATFHPVQSLMSWDSTGPNTGEFCGNPTTGFDNAGY
::*:* * .: * *: ::*::: .* * : ** :
Mflv_2346|M.gilvum_PYR-GCK CYADRTIGWDRGELLPSLQRAHGDIGVVMVLAHEYGHAVSRLSGLSGRD-
Mvan_4299|M.vanbaalenii_PYR-1 CYPDRTIGWDRGELLPGLQRAHGDMGVVMVLAHEYGHAISRLAGLTKKD-
TH_1205|M.thermoresistible__bu CFPERTIGWDRGQMLPRLRRANGDMAVVMVLAHEYGHWVSRQAGLTSRR-
Mb0427|M.bovis_AF2122/97 SSRCNLIAWDRGVFMAVAQEYFGDMSVNGVLAHEFGHALQVMANLVTRK-
Rv0419|M.tuberculosis_H37Rv SSRCNLIAWDRGVFMAVAQEYFGDMSVNGVLAHEFGHALQVMANLVTRK-
MMAR_0728|M.marinum_M TSRCNMIAWDRGVFMPVAQEYFGEISVTGVLAHEFGHALQVMAKLVTRR-
MUL_3589|M.ulcerans_Agy99 TSRCNMIAWDRGVFMPVAQEYFGEISVTGVLAHEFGHALQVMAKLVTRR-
MAV_4736|M.avium_104 TSRCNLIAWDRGVFMPVAQKYFGDMSVTGVLAHEFGHALQQMAALVTKR-
MSMEG_4893|M.smegmatis_MC2_155 CFNDDVMAWDRGVAIPVAAQYFGQMGVVGVMAHEYGHAVQHQAGLIDTRS
MAB_1470c|M.abscessus_ATCC_199 CINQNALGWDRGALLPKLVRMFGTMAVPMTLAHEYGHAVAHQSEMVDSR-
:.**** :. . * :.* .:***:** : : :
Mflv_2346|M.gilvum_PYR-GCK TPTLVAEQQADCFSGAYMRWVAEDSSQRFSLSTGEGLNNVLAGVISFRDP
Mvan_4299|M.vanbaalenii_PYR-1 TPTLVAEQQADCFGGAYMRWVAEDSSARFTLSTGEGLNNVLAGVISFRDP
TH_1205|M.thermoresistible__bu SPILVAEQQADCFAGTYLRWVAEGNSRRFTLSTADGLNDVLAAVVAVRDP
Mb0427|M.bovis_AF2122/97 DPTIVREQQADCFAGVYLWWVAEGKSTRFTLSTADGLDHVLAGIITTRDP
Rv0419|M.tuberculosis_H37Rv DPTIVREQQADCFAGVYLWWVAEGKSTRFTLSTADGLDHVLAGIITTRDP
MMAR_0728|M.marinum_M DPTIVREQQADCFAGVYLYWVAEGKSPRFRLSTADGLDHVLAGIITTRDP
MUL_3589|M.ulcerans_Agy99 DPTIVREQQADCFAGVYLYWVAEGKSPRFRLSTADGLDHVFAGIITTRDP
MAV_4736|M.avium_104 DATIVREQQADCFAGVYLFWVADGKSPRFTLSTADGLDHVLAGIITTRDP
MSMEG_4893|M.smegmatis_MC2_155 TPVLIAEQQADCFAGVYLRWVAAGNSPRFALSTGDGLNHVLAGIIYIRDP
MAB_1470c|M.abscessus_ATCC_199 TQTLVGEQMADCLAGTYLRWVAQNNSPRFTLSTGDGLNKVIAALIAFRDD
:: ** ***:.*.*: *** ..* ** ***.:**:.*:*.:: **
Mflv_2346|M.gilvum_PYR-GCK LLNEGDPDAGYDEHGSAFERVSAFQFGFTDGPSACSGIDLDEIGQRRGDL
Mvan_4299|M.vanbaalenii_PYR-1 LLAENDPDVGFDEHGTAFERLSAFQFGFTDGPSACAAIDPAEIDQRRGEL
TH_1205|M.thermoresistible__bu VLTEGDWRAGLNEHGSAFERLSAFQFGFTNGPSACAAIDMPEIKQRRGEL
Mb0427|M.bovis_AF2122/97 VMEADAE--NDDEHGSALDRVSAFQLGFINGTPACAAIDEDEVERRRGDL
Rv0419|M.tuberculosis_H37Rv VMEADAE--NDDEHGSALDRVSAFQLGFINGTPACAAIDEDEVERRRGDL
MMAR_0728|M.marinum_M VMDTDEQ--NDDEHGSALDRVSAFQMGFLSGTPACAAINESEIVRRRGDL
MUL_3589|M.ulcerans_Agy99 VMDTDEQ--NDDEHGSALDRVSAFQMGLLSGTPACAAINESEIVRRRGDL
MAV_4736|M.avium_104 VMDSETA--NDDAHGSALDRISAFQMGFLNGASACASINRKEIEQRRGDL
MSMEG_4893|M.smegmatis_MC2_155 LMTQLDAALTGNEHGSALDRVSAFQIGFSGNVDQCAAIDMDDIKKRRGDL
MAB_1470c|M.abscessus_ATCC_199 PIEIGDI---VDSHGSAFERVSAFQHGFIDGAAECTRIDMAEITKRRGGL
: : **:*::*:**** *: .. *: *: :: :*** *
Mflv_2346|M.gilvum_PYR-GCK PVLLPE-----DQTGELPVTEDSARSIVDAMEILFEP--QNPPALSFEPA
Mvan_4299|M.vanbaalenii_PYR-1 PVLLPE-----DQTGELPVTEDSTRSIIDAMGILFSP--KNPPALSFDAD
TH_1205|M.thermoresistible__bu PVLLPE-----NQTGELPVTERSVRTLLDALTIVFRP--AAPPRLTLAAA
Mb0427|M.bovis_AF2122/97 PTTLRVDASGNPETGEVGINEETLSTLMELMGKIFSP--KNPPTLSYQPA
Rv0419|M.tuberculosis_H37Rv PTALRVDASGNPETGEVGINEETLSTLMELMGKIFSP--KNPPTLSYQPA
MMAR_0728|M.marinum_M PTGLRVDSSGNPETGEVPINQDTLTTLMELLGKIFSP--TDPPTLSYQQA
MUL_3589|M.ulcerans_Agy99 PTGLRVDSSGNSETGEVPINQDTLTTLMELLGKIFSP--TDPPTLSYQQA
MAV_4736|M.avium_104 PYALRVDTSGRPETGEVPINQDTLSTLMELLGKIFAP--ANPPALSYQPA
MSMEG_4893|M.smegmatis_MC2_155 PKFLDG-VVADPSAGNDEITEDLLTDVMDSLGQIFAP--ADPPSLTIGDA
MAB_1470c|M.abscessus_ATCC_199 PVSPPS-----GVSTQIPISVDTLRTLMTVLKSVYPTNGTTAPRLTVSGA
* : : :. :: : :: . .* *:
Mflv_2346|M.gilvum_PYR-GCK DADNCADARPSPPASFCPATNTIVVDLPALEEMGTQADPDDGTALAAGDN
Mvan_4299|M.vanbaalenii_PYR-1 EAESCADARPSPPASFCPATNTIVVDLPALEEMGAQADPQDGGSLATGDN
TH_1205|M.thermoresistible__bu EAESCPDARPSPPASYCPATNVIAVDLDGLAALGAQSDTTDG--LASGDN
Mb0427|M.bovis_AF2122/97 G---CPDAKPSPPAAYCPATNTIVVDLPALARMGKVASAAE-HSLPQGDD
Rv0419|M.tuberculosis_H37Rv G---CPDAKPSPPAAYCPATNTIVVDLPALARMGKVASAAE-HSLPQGDD
MMAR_0728|M.marinum_M G---CPDAKPSPPASYCPATNTITVDLPALAAIGKVADSKQ-HSLPQGDD
MUL_3589|M.ulcerans_Agy99 G---CPDAKPSPPASYCPATNTITVDLPALAAIGKVADSKQ-HSLPQGDD
MAV_4736|M.avium_104 T---CPDAKPSPPASYCPATNTIAVDLPKLAALGRVADENE-HTLPQGDD
MSMEG_4893|M.smegmatis_MC2_155 SG--CPDAQPTPPASYCPATNTIVVDMPGLQALGETKTENEEQELLQGDN
MAB_1470c|M.abscessus_ATCC_199 ACDEPGRRVASPPAIYCPISNTVALDLRRLTDLAQPTHDQG---QITGDF
.:*** :** :*.:.:*: * :. **
Mflv_2346|M.gilvum_PYR-GCK TAYSVLVSRYMLAIQHERGGVALNSAEAALRTACLTGVATVKMSEEVT-T
Mvan_4299|M.vanbaalenii_PYR-1 MAYSVLVSRYMLAIQQEHGGAALDTAEAALRTACLTGVATVKMTEDVT-T
TH_1205|M.thermoresistible__bu TAYSVLASRFVLALQREHGGVALNDAKAALRTACLTGVATTKLSAGVV-T
Mb0427|M.bovis_AF2122/97 TSLSIVMSRYALAVQHERG-LPMQSPWTALRTACLTGVAHRKMAVPID-L
Rv0419|M.tuberculosis_H37Rv TSLSIVMSRYALAVQHERG-LPMQSPWTALRTACLTGVAHRKMAVPID-L
MMAR_0728|M.marinum_M TALSIVMSRYALSVQHQRD-LPMQSPWTALRTACLTGVVHREMAEPIE-L
MUL_3589|M.ulcerans_Agy99 TALSIVMSRYALSVQHQRD-LPMQSPWTALRTACLTGVVHREMAEPIE-L
MAV_4736|M.avium_104 TALSVVMSRYALAVQHERG-LAMQSPWTALRTACLTGVVHRRMAEPIE-L
MSMEG_4893|M.smegmatis_MC2_155 SAISVLTSRYALAVAREKG-HAIDNPVAAMRTACLTGVAQAAMAEP----
MAB_1470c|M.abscessus_ATCC_199 TAFSVVISRYALAVENQHG-LPLDNGQSALRAACLTGVASAALAKPTTPT
: *:: **: *:: .::. .:: :*:*:******. ::
Mflv_2346|M.gilvum_PYR-GCK PDGDTIALTAGDVDEAVSGMLTNGLAASDVNGESVPSGFSRIDAFRVGVL
Mvan_4299|M.vanbaalenii_PYR-1 PDGDTIALTAGDIDEAVSGILTNGLVASDVNGASVPSGFSRIDAFRLGVL
TH_1205|M.thermoresistible__bu SDGTSISLTAGDLDEAVSGMLTNGLVASDVHGETVPSGFSRIDAYRLGVL
Mb0427|M.bovis_AF2122/97 PSGQQLVLTAGDLDEAVSGLLTNRMVASDADGVSVPAGFTRIAAFRAGVG
Rv0419|M.tuberculosis_H37Rv PSGQQLVLTAGDLDEAVSGLLTNRMVASDADGVSVPAGFTRIAAFRAGVG
MMAR_0728|M.marinum_M PSGQQLVLTAGDLDEAVSGLLTNRMVASDADGISVPAGFTRIAAFRAGVS
MUL_3589|M.ulcerans_Agy99 PSGQQLVLTAGDLDEAVSGLLTNRMVASDADGISVPAGFTRIATFRAGVS
MAV_4736|M.avium_104 PSHNQLLLTAGDLDEAVAGLLTNHLVASDADGTSVPAGFTRIAAFRGGVT
MSMEG_4893|M.smegmatis_MC2_155 --GLPVRLSAGDTDEAISGLLTNGLAASDVNGALLPAGFTRILGYRSGLQ
MAB_1470c|M.abscessus_ATCC_199 QEPNPVVLSAGDLDEAVSGLLTNGMVASDVNGVTIPAGFSRIDAFRDGVA
: *:*** ***::*:*** :.***..* :*:**:** :* *:
Mflv_2346|M.gilvum_PYR-GCK GDADRCFTRFP-
Mvan_4299|M.vanbaalenii_PYR-1 SDTERCFKRFP-
TH_1205|M.thermoresistible__bu GDASRCYQRFG-
Mb0427|M.bovis_AF2122/97 GDMDACYARYPG
Rv0419|M.tuberculosis_H37Rv GDMDACYARYPG
MMAR_0728|M.marinum_M GDMEACYSRYPG
MUL_3589|M.ulcerans_Agy99 GDMEACYSRYPG
MAV_4736|M.avium_104 GNADACYSRYPG
MSMEG_4893|M.smegmatis_MC2_155 GDDAQCYQRFP-
MAB_1470c|M.abscessus_ATCC_199 GTTDRCLTRYG-
. * *: