For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SAAAVLLVAGCSTFVEGRALSMLNDPFLVGGLPATNGPSGIRSNAPAPTGKVLNTDNGSIDSLSLLSIND IEDYWRSVYGQSLKGKFVPISKIVSYNSTDPSSPIVCHNDTYQLVNAFYTSRCNLIAWDRGVFMPVAQKY FGDMSVTGVLAHEFGHALQQMAALVTKRDATIVREQQADCFAGVYLFWVADGKSPRFTLSTADGLDHVLA GIITTRDPVMDSETANDDAHGSALDRISAFQMGFLNGASACASINRKEIEQRRGDLPYALRVDTSGRPET GEVPINQDTLSTLMELLGKIFAPANPPALSYQPATCPDAKPSPPASYCPATNTIAVDLPKLAALGRVADE NEHTLPQGDDTALSVVMSRYALAVQHERGLAMQSPWTALRTACLTGVVHRRMAEPIELPSHNQLLLTAGD LDEAVAGLLTNHLVASDADGTSVPAGFTRIAAFRGGVTGNADACYSRYPG*
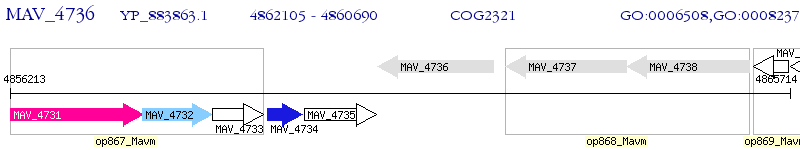
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4736 | - | - | 100% (471) | LpqM protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0427 | lpqM | 0.0 | 78.21% (468) | lipoprotein peptidase LpqM |
| M. gilvum PYR-GCK | Mflv_2346 | - | 1e-132 | 49.27% (477) | LpqM |
| M. tuberculosis H37Rv | Rv0419 | lpqM | 0.0 | 78.42% (468) | lipoprotein peptidase LpqM |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1470c | - | 1e-107 | 44.07% (481) | lipoprotein peptidase LpqM |
| M. marinum M | MMAR_0728 | lpqM | 0.0 | 79.23% (467) | lipoprotein peptidase LpqM |
| M. smegmatis MC2 155 | MSMEG_4893 | - | 1e-135 | 53.99% (476) | putative neutral zinc metallopeptidase |
| M. thermoresistible (build 8) | TH_2655 | - | 1e-123 | 50.53% (475) | PUTATIVE MscS Mechanosensitive ion channel |
| M. ulcerans Agy99 | MUL_3589 | lpqM | 0.0 | 78.16% (467) | lipoprotein peptidase LpqM |
| M. vanbaalenii PYR-1 | Mvan_4299 | - | 1e-128 | 48.53% (476) | LpqM |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2346|M.gilvum_PYR-GCK -----------MSPSRSSKPSRFRSSAALLAGTAVLAGA-ACST--MLQG
Mvan_4299|M.vanbaalenii_PYR-1 -----------MSPSRRPG-TRIGVAAAVCA---VLAAT-ACST--TLQG
Mb0427|M.bovis_AF2122/97 MHGRGRYRPLVRCVRPRRVAASVRTPIACLAAVVVIAG---CTT--VVDG
Rv0419|M.tuberculosis_H37Rv MHGRGRYRPLVRCVRPRRVAASVRTPIACLAAVVVIAG---CTT--VVDG
MMAR_0728|M.marinum_M MDNRGRKWLGVRRSRHGGLIASLWAATAFLTALLVVAG---CTT--IVDG
MUL_3589|M.ulcerans_Agy99 MDNRGRKWLGVRRSRHSGLIASLWAATAFLTALLVVAG---CTT--IADG
MAV_4736|M.avium_104 ---------------------------MSAAAVLLVAG---CST--FVEG
MSMEG_4893|M.smegmatis_MC2_155 ------------------MVARVCWGAFLAVCAITLAG---CSSDPVIAG
TH_2655|M.thermoresistible__bu -----MPTTATLRSAQRGAVRWARIPVALAVVVLCVLAGTGCGQRYVLDG
MAB_1470c|M.abscessus_ATCC_199 --------------MSIPLRWFHTSTTILACCVLGLAG---CAN--TVDG
: . * *
Mflv_2346|M.gilvum_PYR-GCK NAVSVFDDPFKVAGMPATDGPTGLRPDAESPARDVTGTDGGDIDNLAASA
Mvan_4299|M.vanbaalenii_PYR-1 NAVSVFDDPFRVAGMPATDGPTGVRPNAEPPARDVEGTDGGAIDELAASA
Mb0427|M.bovis_AF2122/97 RALSILNDPFRVGGLPATNGPSGARPDAPAASGTVINTNNGAIDKLSLLS
Rv0419|M.tuberculosis_H37Rv RALSILNDPFRVGGLPATNGPSGARPDAPAASGTVINTNNGAIDKLSLLS
MMAR_0728|M.marinum_M RALSILNDPFRVGGLPATNGPSGPRPDGPPPTGTVINTNNGPIDKLSLLS
MUL_3589|M.ulcerans_Agy99 RALSILNDPFRVGGLPATNGPSGPRPDGPPPTGTVINTNNGPIDKLSLLS
MAV_4736|M.avium_104 RALSMLNDPFLVGGLPATNGPSGIRSNAPAPTGKVLNTDNGSIDSLSLLS
MSMEG_4893|M.smegmatis_MC2_155 RATSMLFVPDRVGGLQVTEGPSGTRPDAPAPRRQAENTDGGQIDNLALLA
TH_2655|M.thermoresistible__bu RAVSMRYDPFRVAGLPTTDGPSGPRVPLPGSTARVTNSDGGEIDRSALLA
MAB_1470c|M.abscessus_ATCC_199 TPVSQLGNAYQIAGLPVVDGPSGLRPHPTSADRAVRGTDSGQMDRLAVNA
. * . :.*: ..:**:* * . . .::.* :* : :
Mflv_2346|M.gilvum_PYR-GCK VSDIEDYWATAYE-DAFDERFKPVAELISWDANGFDG-EFCGDTTYGL-V
Mvan_4299|M.vanbaalenii_PYR-1 ISDIEDYWAVAYE-ETFDEKFRPVAELISWDANGFDG-EFCGEDTYGL-I
Mb0427|M.bovis_AF2122/97 VNDIEDYWMAVYS-ESLKGTFRPVGKLVSYDSNDPSSPIVCHIDTYQL-V
Rv0419|M.tuberculosis_H37Rv VNDIEDYWMAVYS-ESLKGTFRPVGKLVSYDSNDPSSPIVCHIDTYQL-V
MMAR_0728|M.marinum_M VNDIQDYWKANY--EPLQGTFRPVSKLVSYDSNDPNSPIVCHNDTYEL-V
MUL_3589|M.ulcerans_Agy99 VNDIQDYWKANY--EPLQGTFRPVSKLVSYDSNDPNSPIVCHNDTYEL-V
MAV_4736|M.avium_104 INDIEDYWRSVYG-QSLKGKFVPISKIVSYNSTDPSSPIVCHNDTYQL-V
MSMEG_4893|M.smegmatis_MC2_155 VDDIEDFWSQNYSGGSLRGSFTPIRRLVSYNSDQSPGIEICGEDTEGL-T
TH_2655|M.thermoresistible__bu IEDIEDFWSQHYP-EAFSGTFAPVQTLVSIDPSDPSSPAVCGAEAAELRY
MAB_1470c|M.abscessus_ATCC_199 IADIEDFWQEAYP-KTFWATFHPVQSLMSWDSTGPNTGEFCGNPTTGF-D
: **:*:* * .: * *: ::* :. .* : :
Mflv_2346|M.gilvum_PYR-GCK NAGYCYADRTIGWDRGELLPSLQRAHGDIGVVMVLAHEYGHAVSRLSGLS
Mvan_4299|M.vanbaalenii_PYR-1 NAGYCYPDRTIGWDRGELLPGLQRAHGDMGVVMVLAHEYGHAISRLAGLT
Mb0427|M.bovis_AF2122/97 NAFFSSRCNLIAWDRGVFMAVAQEYFGDMSVNGVLAHEFGHALQVMANLV
Rv0419|M.tuberculosis_H37Rv NAFFSSRCNLIAWDRGVFMAVAQEYFGDMSVNGVLAHEFGHALQVMANLV
MMAR_0728|M.marinum_M NAFFTSRCNMIAWDRGVFMPVAQEYFGEISVTGVLAHEFGHALQVMAKLV
MUL_3589|M.ulcerans_Agy99 NAFFTSRCNMIAWDRGVFMPVAQEYFGEISVTGVLAHEFGHALQVMAKLV
MAV_4736|M.avium_104 NAFYTSRCNLIAWDRGVFMPVAQKYFGDMSVTGVLAHEFGHALQQMAALV
MSMEG_4893|M.smegmatis_MC2_155 NAFYCFNDDVMAWDRGVAIPVAAQYFGQMGVVGVMAHEYGHAVQHQAGLI
TH_2655|M.thermoresistible__bu NAAYCRTRDVLVWDREYLLPVANTFFGVMAVNGVLAHEYGHAVQVQARLV
MAB_1470c|M.abscessus_ATCC_199 NAGYCINQNALGWDRGALLPKLVRMFGTMAVPMTLAHEYGHAVAHQSEMV
** : : *** :. .* :.* .:***:***: : :
Mflv_2346|M.gilvum_PYR-GCK GRD-TPTLVAEQQADCFSGAYMRWVAEDSSQRFSLSTGEGLNNVLAGVIS
Mvan_4299|M.vanbaalenii_PYR-1 KKD-TPTLVAEQQADCFGGAYMRWVAEDSSARFTLSTGEGLNNVLAGVIS
Mb0427|M.bovis_AF2122/97 TRK-DPTIVREQQADCFAGVYLWWVAEGKSTRFTLSTADGLDHVLAGIIT
Rv0419|M.tuberculosis_H37Rv TRK-DPTIVREQQADCFAGVYLWWVAEGKSTRFTLSTADGLDHVLAGIIT
MMAR_0728|M.marinum_M TRR-DPTIVREQQADCFAGVYLYWVAEGKSPRFRLSTADGLDHVLAGIIT
MUL_3589|M.ulcerans_Agy99 TRR-DPTIVREQQADCFAGVYLYWVAEGKSPRFRLSTADGLDHVFAGIIT
MAV_4736|M.avium_104 TKR-DATIVREQQADCFAGVYLFWVADGKSPRFTLSTADGLDHVLAGIIT
MSMEG_4893|M.smegmatis_MC2_155 DTRSTPVLIAEQQADCFAGVYLRWVAAGNSPRFALSTGDGLNHVLAGIIY
TH_2655|M.thermoresistible__bu DAD-TPTLIGEQQADCLAGTYLRWVAEGHSPRFTLNTTGDLDRVIAGAIA
MAB_1470c|M.abscessus_ATCC_199 DSR-TQTLVGEQMADCLAGTYLRWVAQNNSPRFTLSTGDGLNKVIAALIA
.:: ** ***:.*.*: *** . * ** *.* .*:.*:*. *
Mflv_2346|M.gilvum_PYR-GCK FRDPLLNEGDPDAGYDEHGSAFERVSAFQFGFTDGPSACSGIDLDEIGQR
Mvan_4299|M.vanbaalenii_PYR-1 FRDPLLAENDPDVGFDEHGTAFERLSAFQFGFTDGPSACAAIDPAEIDQR
Mb0427|M.bovis_AF2122/97 TRDPVMEADAE--NDDEHGSALDRVSAFQLGFINGTPACAAIDEDEVERR
Rv0419|M.tuberculosis_H37Rv TRDPVMEADAE--NDDEHGSALDRVSAFQLGFINGTPACAAIDEDEVERR
MMAR_0728|M.marinum_M TRDPVMDTDEQ--NDDEHGSALDRVSAFQMGFLSGTPACAAINESEIVRR
MUL_3589|M.ulcerans_Agy99 TRDPVMDTDEQ--NDDEHGSALDRVSAFQMGLLSGTPACAAINESEIVRR
MAV_4736|M.avium_104 TRDPVMDSETA--NDDAHGSALDRISAFQMGFLNGASACASINRKEIEQR
MSMEG_4893|M.smegmatis_MC2_155 IRDPLMTQLDAALTGNEHGSALDRVSAFQIGFSGNVDQCAAIDMDDIKKR
TH_2655|M.thermoresistible__bu MRDPISVVRLLVPPSEAHGTALDRVSALQQGFDVGAEACAAIDFDDIEQR
MAB_1470c|M.abscessus_ATCC_199 FRDDPIEIGDI---VDSHGSAFERVSAFQHGFIDGAAECTRIDMAEITKR
** : **:*::*:**:* *: . *: *: :: :*
Mflv_2346|M.gilvum_PYR-GCK RGDLPVLLPE-----DQTGELPVTEDSARSIVDAMEILFEP--QNPPALS
Mvan_4299|M.vanbaalenii_PYR-1 RGELPVLLPE-----DQTGELPVTEDSTRSIIDAMGILFSP--KNPPALS
Mb0427|M.bovis_AF2122/97 RGDLPTTLRVDASGNPETGEVGINEETLSTLMELMGKIFSP--KNPPTLS
Rv0419|M.tuberculosis_H37Rv RGDLPTALRVDASGNPETGEVGINEETLSTLMELMGKIFSP--KNPPTLS
MMAR_0728|M.marinum_M RGDLPTGLRVDSSGNPETGEVPINQDTLTTLMELLGKIFSP--TDPPTLS
MUL_3589|M.ulcerans_Agy99 RGDLPTGLRVDSSGNSETGEVPINQDTLTTLMELLGKIFSP--TDPPTLS
MAV_4736|M.avium_104 RGDLPYALRVDTSGRPETGEVPINQDTLSTLMELLGKIFAP--ANPPALS
MSMEG_4893|M.smegmatis_MC2_155 RGDLPKFLDG-VVADPSAGNDEITEDLLTDVMDSLGQIFAP--ADPPSLT
TH_2655|M.thermoresistible__bu RGELSSLLFH--SGIPAS-DLRITENTVKSLMEVLDVIFAP--AAPPALE
MAB_1470c|M.abscessus_ATCC_199 RGGLPVSPPS-----GVSTQIPISVDTLRTLMTVLKSVYPTNGTTAPRLT
** *. : : :. : :: : :: . .* *
Mflv_2346|M.gilvum_PYR-GCK FEPADADNCADARPSPPASFCPATNTIVVDLPALEEMGTQADPDDGTALA
Mvan_4299|M.vanbaalenii_PYR-1 FDADEAESCADARPSPPASFCPATNTIVVDLPALEEMGAQADPQDGGSLA
Mb0427|M.bovis_AF2122/97 YQ--PAG-CPDAKPSPPAAYCPATNTIVVDLPALARMGKVASAAE-HSLP
Rv0419|M.tuberculosis_H37Rv YQ--PAG-CPDAKPSPPAAYCPATNTIVVDLPALARMGKVASAAE-HSLP
MMAR_0728|M.marinum_M YQ--QAG-CPDAKPSPPASYCPATNTITVDLPALAAIGKVADSKQ-HSLP
MUL_3589|M.ulcerans_Agy99 YQ--QAG-CPDAKPSPPASYCPATNTITVDLPALAAIGKVADSKQ-HSLP
MAV_4736|M.avium_104 YQ--PAT-CPDAKPSPPASYCPATNTIAVDLPKLAALGRVADENE-HTLP
MSMEG_4893|M.smegmatis_MC2_155 IG--DASGCPDAQPTPPASYCPATNTIVVDMPGLQALGETKTENEEQELL
TH_2655|M.thermoresistible__bu VD--QSG-CPLPDG-LPARYCPDSNTVSVDLAALRRIGTPADESQ-QVLL
MAB_1470c|M.abscessus_ATCC_199 VSGAACDEPGRRVASPPAIYCPISNTVALDLRRLTDLAQPTHDQG---QI
. ** :** :**: :*: * :.
Mflv_2346|M.gilvum_PYR-GCK AGDNTAYSVLVSRYMLAIQHERGGVALNSAEAALRTACLTGVATVKMSEE
Mvan_4299|M.vanbaalenii_PYR-1 TGDNMAYSVLVSRYMLAIQQEHGGAALDTAEAALRTACLTGVATVKMTED
Mb0427|M.bovis_AF2122/97 QGDDTSLSIVMSRYALAVQHERG-LPMQSPWTALRTACLTGVAHRKMAVP
Rv0419|M.tuberculosis_H37Rv QGDDTSLSIVMSRYALAVQHERG-LPMQSPWTALRTACLTGVAHRKMAVP
MMAR_0728|M.marinum_M QGDDTALSIVMSRYALSVQHQRD-LPMQSPWTALRTACLTGVVHREMAEP
MUL_3589|M.ulcerans_Agy99 QGDDTALSIVMSRYALSVQHQRD-LPMQSPWTALRTACLTGVVHREMAEP
MAV_4736|M.avium_104 QGDDTALSVVMSRYALAVQHERG-LAMQSPWTALRTACLTGVVHRRMAEP
MSMEG_4893|M.smegmatis_MC2_155 QGDNSAISVLTSRYALAVAREKG-HAIDNPVAAMRTACLTGVAQAAMAEP
TH_2655|M.thermoresistible__bu QGDNTALSVVVSRYVLAVQQERG-VGLESGVAAMRTACLTGVAQHQMAER
MAB_1470c|M.abscessus_ATCC_199 TGDFTAFSVVISRYALAVENQHG-LPLDNGQSALRAACLTGVASAALAKP
** : *:: *** *:: .::. ::. :*:*:******. ::
Mflv_2346|M.gilvum_PYR-GCK VT-TPDGDTIALTAGDVDEAVSGMLTNGLAASDVNGESVPSGFSRIDAFR
Mvan_4299|M.vanbaalenii_PYR-1 VT-TPDGDTIALTAGDIDEAVSGILTNGLVASDVNGASVPSGFSRIDAFR
Mb0427|M.bovis_AF2122/97 ID-LPSGQQLVLTAGDLDEAVSGLLTNRMVASDADGVSVPAGFTRIAAFR
Rv0419|M.tuberculosis_H37Rv ID-LPSGQQLVLTAGDLDEAVSGLLTNRMVASDADGVSVPAGFTRIAAFR
MMAR_0728|M.marinum_M IE-LPSGQQLVLTAGDLDEAVSGLLTNRMVASDADGISVPAGFTRIAAFR
MUL_3589|M.ulcerans_Agy99 IE-LPSGQQLVLTAGDLDEAVSGLLTNRMVASDADGISVPAGFTRIATFR
MAV_4736|M.avium_104 IE-LPSHNQLLLTAGDLDEAVAGLLTNHLVASDADGTSVPAGFTRIAAFR
MSMEG_4893|M.smegmatis_MC2_155 ------GLPVRLSAGDTDEAISGLLTNGLAASDVNGALLPAGFTRILGYR
TH_2655|M.thermoresistible__bu -----NDLALTLGTGDLDEAVSGILTNGVVASDVNGGVVPSGFTRISAFR
MAB_1470c|M.abscessus_ATCC_199 TTPTQEPNPVVLSAGDLDEAVSGLLTNGMVASDVNGVTIPAGFSRIDAFR
: * :** ***::*:*** :.***.:* :*:**:** :*
Mflv_2346|M.gilvum_PYR-GCK VGVLGDADRCFTRFP-
Mvan_4299|M.vanbaalenii_PYR-1 LGVLSDTERCFKRFP-
Mb0427|M.bovis_AF2122/97 AGVGGDMDACYARYPG
Rv0419|M.tuberculosis_H37Rv AGVGGDMDACYARYPG
MMAR_0728|M.marinum_M AGVSGDMEACYSRYPG
MUL_3589|M.ulcerans_Agy99 AGVSGDMEACYSRYPG
MAV_4736|M.avium_104 GGVTGNADACYSRYPG
MSMEG_4893|M.smegmatis_MC2_155 SGLQGDDAQCYQRFP-
TH_2655|M.thermoresistible__bu SGLAGDVDDCFRRFP-
MAB_1470c|M.abscessus_ATCC_199 DGVAGTTDRCLTRYG-
*: . * *: