For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VMFSRNGIRALRIWTAVVLVFLYIPLLLVLINAFNASRTFAFPPTGFTLAWWADAARSPGMWHALANSVV VGLAATAIALVLGTMIAFAVQRHRFFGRTTVSFLVVLPITLPGIVTGIALNATFTSALGLMLGMATVIIG HATFCIVIVFNNTQARLRQMGSSLQDASADLGASSWQTFRFVTFPMMRGALLAGAILAFALSFDEIVVTT FTAGPTVQTLPIWIFSNLFRPNQAPVINVVAAALTVLAIIPVWLAGRFGGDPAGARV
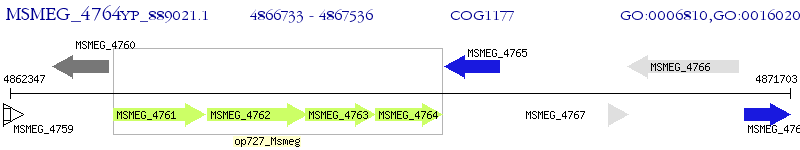
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4764 | - | - | 100% (267) | ABC transporter permease protein |
| M. smegmatis MC2 155 | MSMEG_3277 | - | 2e-38 | 36.44% (247) | polyamine ABC transporter integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_0658 | - | 3e-22 | 28.74% (247) | polyamine ABC-transporter, inner membrane subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2420c | cysW | 1e-14 | 23.69% (249) | sulfate-transport integral membrane protein ABC transporter |
| M. gilvum PYR-GCK | Mflv_3135 | - | 1e-37 | 35.27% (241) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv2398c | cysW | 2e-15 | 24.10% (249) | sulfate-transport integral membrane protein ABC transporter |
| M. leprae Br4923 | MLBr_02093 | pstA1 | 4e-15 | 23.68% (266) | membrane-bound component of phosphate transport |
| M. abscessus ATCC 19977 | MAB_1504 | - | 3e-18 | 26.52% (264) | ABC transporter permease |
| M. marinum M | MMAR_3717 | cysT | 9e-15 | 29.27% (205) | sulfate-transport integral membrane protein ABC transporter |
| M. avium 104 | MAV_1049 | pstA | 2e-14 | 24.66% (223) | phosphate ABC transporter, permease protein PstA |
| M. thermoresistible (build 8) | TH_2990 | - | 1e-14 | 25.30% (249) | PUTATIVE sulfate ABC transporter, permease protein CysW |
| M. ulcerans Agy99 | MUL_3660 | cysT | 6e-15 | 29.27% (205) | sulfate-transport integral membrane protein ABC transporter |
| M. vanbaalenii PYR-1 | Mvan_3390 | - | 8e-37 | 35.10% (245) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3135|M.gilvum_PYR-GCK --MTTQAGAALATAAQQDPAPKKVRTGPNWGDLLLRVAAGLVLLYLFLPI
Mvan_3390|M.vanbaalenii_PYR-1 --MTTQAGAAIATAAQHDPKPKKVRGRTNWGDLALKMVAGLVLLYLFLPI
MSMEG_4764|M.smegmatis_MC2_155 ------------------------MMFSRNGIRALRIWTAVVLVFLYIPL
Mb2420c|M.bovis_AF2122/97 ----------------------------------MTSLPAARYLVRSVAL
Rv2398c|M.tuberculosis_H37Rv ----------------------------------MTSLPAARYLVRSVAL
TH_2990|M.thermoresistible__bu -------------------------VTSGSNGSGLTPARPARYLLRFVAL
MMAR_3717|M.marinum_M MTVTAHSGQSDPPAVAPDPGQRADGGPQWISGLGRSGRRGATALQVGVSM
MUL_3660|M.ulcerans_Agy99 MTVTAHSGQSDPPAVAPDLGQRADGGPQWISGPGRSGRRGATALQVGVSM
MLBr_02093|M.leprae_Br4923 ----MN--LDTLDRPVKAVVLRPISVRRRIKNNVATMFFFTSFLIALVPL
MAV_1049|M.avium_104 ----MTRAVDALDRPVKTEVFRPLSVRRRITNNAATIFFLGSFVVALVPL
MAB_1504|M.abscessus_ATCC_1997 -------------------------MLLWNGRSRLILWSVFAVVITVVFI
: : :
Mflv_3135|M.gilvum_PYR-GCK FVIILFSFN-KPAGKFNYSWQGFTLDNWLDPFKYPALTEALKLSLNVAAV
Mvan_3390|M.vanbaalenii_PYR-1 FVIVLFSFN-KPAGKFNYTWQGFTLENWLDPFKYPPLTEALKLSLNVAAV
MSMEG_4764|M.smegmatis_MC2_155 LLVLINAFN-ASR-TFAFPPTGFTLAWWADAARSPGMWHALANSVVVGLA
Mb2420c|M.bovis_AF2122/97 GYVFVLLI--VPVALILWRTFEPGFGQFYAWISTPAAISALNLSLLVVAI
Rv2398c|M.tuberculosis_H37Rv GYVFVLLI--VPVALILWRTFEPGFGQFYAWISTPAAISALNLSLLVVAI
TH_2990|M.thermoresistible__bu SYVVVLVA--VPVGLILWRTFAPGLGEFIASVTTPAAISALQLSLLVVAI
MMAR_3717|M.marinum_M VWLSVIVL--LPLAAIAWQAADGGWHAFWLSVSSHAALESLRVTLTVSAG
MUL_3660|M.ulcerans_Agy99 VWLSVIVL--LPLAAIAWQAADGGWHAFWLSVSSHAALESLRVTLTVSAG
MLBr_02093|M.leprae_Br4923 VWLLSVVVARGFYAVTRFDWWTHSLRGVMPEEFAGGVYHALYGSLVQAVV
MAV_1049|M.avium_104 IWVLSVVLERGWYAVTRSGWWTHSLHGVLPEQFAGGVYHALYGTVVQAGV
MAB_1504|M.abscessus_ATCC_1997 APIATVVLAGFSGAWTGPLPTQLSFMRYEKALG-GDDLASMAVSLQTAVM
: :: ::
Mflv_3135|M.gilvum_PYR-GCK STAVALVLGTLVAIALVRQRWR-GQRAVDTFLILPLTAPEVVMGAALLTL
Mvan_3390|M.vanbaalenii_PYR-1 STAVALVLGTLVAIALVRQRWR-GQRAVDTFLVIPLTAPEVVMGAALLTL
MSMEG_4764|M.smegmatis_MC2_155 ATAIALVLGTMIAFAVQRHRFF-GRTTVSFLVVLPITLPGIVTGIALNAT
Mb2420c|M.bovis_AF2122/97 VVPLNVIFGVTTALVLARNRFR-GKGVLQAIIDLPFAVSPVIVGVSLILL
Rv2398c|M.tuberculosis_H37Rv VVPLNVIFGVTTALVLARNRFR-GKGVLQAIIDLPFAVSPVIVGVSLILL
TH_2990|M.thermoresistible__bu VVPLNVVFGVPTALVLARNRFR-GKAALQAVIDLPFAVSPIIVGVALILL
MMAR_3717|M.marinum_M VTLLNTVFGLLIAWVLIRDEFV-GKRLVDAVIDLPFALPTIVASLVMLAL
MUL_3660|M.ulcerans_Agy99 VTLLNAVFGLLIAWGLIRDEFV-GKRLVDAVIDLPFALPTIVASLVMLAL
MLBr_02093|M.leprae_Br4923 AAIMAVPLGSMTAVYLVEYGSGPFVRVTTFMVDVLAGVPSIVAALFIFCL
MAV_1049|M.avium_104 AAAMAVPLGLMTAVFLVEYGSGRLVRLTTFMVDVLAGVPSIVAALFIFSL
MAB_1504|M.abscessus_ATCC_1997 SSGVALLLGTWAALSVREVPSW-LRRVIDAVFHLPIAIPSVAIGLGLLTA
: :* * : . .. : . : . :
Mflv_3135|M.gilvum_PYR-GCK FLDFN------------TAAGYTTIVLSHIAFEVSFIAMTVRARVRGFDW
Mvan_3390|M.vanbaalenii_PYR-1 FLDLS------------WPAGYTTILLSHIAFEVSFIAMTVRARVRGFDW
MSMEG_4764|M.smegmatis_MC2_155 FTSALG-----------LMLGMATVIIGHATFCIVIVFNNTQARLRQMGS
Mb2420c|M.bovis_AF2122/97 WGSAGALGFVEQDLGFKIIFGLPGIVLASMFVTCPFVVREVEPVLHELGT
Rv2398c|M.tuberculosis_H37Rv WGSAGALGFVEQDLGFKIIFGLPGIVLGSMFVTCPFVVREVEPVLHELGT
TH_2990|M.thermoresistible__bu WGTAGFLGFVENSLGLKIIFGFPGIVLASIFVTVPFVIREVEPVLHEIGT
MMAR_3717|M.marinum_M YGHN-------SPLGLHLQHTVWGVGVALAFVTLPFVVRAVQPVLLELDR
MUL_3660|M.ulcerans_Agy99 YGHN-------SPLGLHLQHTVWGVGVALAFVTLPFVVRAVQPVLLELDR
MLBr_02093|M.leprae_Br4923 WIATLG-----------FQQSAFAVSLALVLLMLPVVVRSTEEILRLVPD
MAV_1049|M.avium_104 WIATLG-----------FQQSSFAVSLALVLLMLPVVVRSAEEMLRLVPD
MAB_1504|M.abscessus_ATCC_1997 FNEPP----------LLLGGTKWIVIVAHTALVLAFAFSSVSAALERLDP
: : :. . . . : .
Mflv_3135|M.gilvum_PYR-GCK TLEDASLDLGASPTRTFFKVTLPLIVPGIVAAAMLSFALSLDDFIITYFV
Mvan_3390|M.vanbaalenii_PYR-1 TLEDASLDLGAGPARTFFKVTLPLIVPGIVAAAMLSFALSLDDFIITYFV
MSMEG_4764|M.smegmatis_MC2_155 SLQDASADLGASSWQTFRFVTFPMMRGALLAGAILAFALSFDEIVVTTFT
Mb2420c|M.bovis_AF2122/97 DQEQAAATLGSGWWQTFWRITLPSIRWGLTYGIVLTVARTLGEYGAVIIV
Rv2398c|M.tuberculosis_H37Rv DQEQAAATLGSGWWQTFWRITLPSIRWGLTYGIVLTVARTLGEYGAVIIV
TH_2990|M.thermoresistible__bu DQEEAAATLGATWWQTFWRVTLPSIRWGLTYGVVLTVARTLGEFGAVLMV
MMAR_3717|M.marinum_M ETEEAAASLGATGWKVFTSVIFPSLMPALLSGAGLAFSRAIGEFGSVVLI
MUL_3660|M.ulcerans_Agy99 ETEEAAASLGATGWKVFTSVIFPSLMPALLSGAGLAFSRAIGEFGSVVLI
MLBr_02093|M.leprae_Br4923 ELREACYALGIPKWKTIVRIVFPIATPGIISGVLLSIARVIGETAPVLVL
MAV_1049|M.avium_104 DLREASYALGVPQWKTIVHIVFPIAMPGIVSGILLSIARVIGETAPVLVL
MAB_1504|M.abscessus_ATCC_1997 AYRQVAESLGAGSVRVVRTVTLPLLMPSLGAAAGLAIALSMGELGATVMV
.:.. ** :.. : :* .: . *:.: :.: . .
Mflv_3135|M.gilvum_PYR-GCK SG------------SEVTYPLYVN-AAVKAA--VPPQINVLATAILVVSL
Mvan_3390|M.vanbaalenii_PYR-1 SG------------AEVTYPLYVN-AAVKAA--VPPQINVLATAILVVSL
MSMEG_4764|M.smegmatis_MC2_155 AGP-----------TVQTLPIWIFSNLFRPN--QAPVINVVAAALTVLAI
Mb2420c|M.bovis_AF2122/97 SSNL--------PGTSQTLTLLVSDRYHRGAEYGAYALSTLLMAVSVVVL
Rv2398c|M.tuberculosis_H37Rv SSNL--------PGTSQTLTLLVSDRYHRGAEYGAYALSTLLMAVSVVVL
TH_2990|M.thermoresistible__bu SSNL--------PGVSQTLTLLVHDRYNRGAEYGAYAISTLLMAVAVVVL
MMAR_3717|M.marinum_M GGAV--------PGETEVSSQWIRTLIENDDRTGAAAISIVLLSISFAVL
MUL_3660|M.ulcerans_Agy99 GGAV--------PGETEVSSQWIRTLIENDDRTGAAAISIVLLSISFAVL
MLBr_02093|M.leprae_Br4923 VGYSRSINFDIFDGNMASLPLLIYTELTNPEYAGFLRVWGAALTLIILVA
MAV_1049|M.avium_104 VGYSRSINLDIFHGNMASLPLLIYTELTNPEHAGFLRIWGAALTLIIIVA
MAB_1504|M.abscessus_ATCC_1997 YPSS-----------WRTLPVTIFGLSDRGQVFSAAASTTLLLMVTLLAL
. : . : .
Mflv_3135|M.gilvum_PYR-GCK LLLAIGTLYRRKKIDI-----
Mvan_3390|M.vanbaalenii_PYR-1 VLLAVSTLYRRKRFAG-----
MSMEG_4764|M.smegmatis_MC2_155 IPVWLAGRFGGDPAGARV---
Mb2420c|M.bovis_AF2122/97 IVQMVLDAHRARAVSEG----
Rv2398c|M.tuberculosis_H37Rv IVQMVLDARRARAVSEG----
TH_2990|M.thermoresistible__bu IFQVILDARRAKSNQ------
MMAR_3717|M.marinum_M LVLRVLGARAAKREELAP---
MUL_3660|M.ulcerans_Agy99 LVLRVLGARAAKREELAP---
MLBr_02093|M.leprae_Br4923 GINLFGAAFRFLATRRCSGCR
MAV_1049|M.avium_104 VINVVAAATRFLAGRRR----
MAB_1504|M.abscessus_ATCC_1997 LVVGRLRGKVAFR--------