For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAAGQGLTDRARLAFLLTPPLAWLVVAYLGSLAVLLVSAFWQTNEFTGAVVRAFTLHNIERVLTDEVFRV ATLRTLGVALTVTVICAALAVPIALYMAKIASPRARLILVVAVTTPLWASYLVKAYAWRMLLSPEGPLSW ATGFTPGYGIVATVATLTYLWLPYMVIPVYAAFERIPDALVDASSDLGASDFATMRMVLAPLVFPGIAAG SIFTFSLSLGDYIAVTIVGGKTQMLGNIIYGQLVTANNQPLAAALSIIPLAAIVAYLLAMRRTGALENV
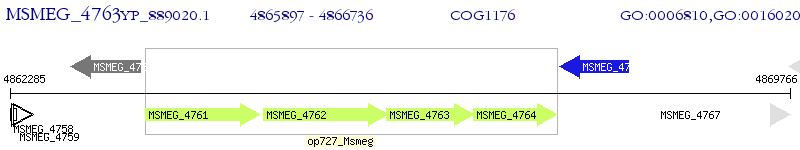
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4763 | - | - | 100% (279) | ABC transporter permease protein |
| M. smegmatis MC2 155 | MSMEG_3279 | - | 1e-29 | 32.42% (219) | polyamine ABC transporter permease protein |
| M. smegmatis MC2 155 | MSMEG_0659 | - | 2e-25 | 27.01% (274) | polyamine ABC transporter permease protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1889 | modB | 2e-14 | 30.33% (211) | molbdenum-transport integral membrane protein ABC MODB |
| M. gilvum PYR-GCK | Mflv_3136 | - | 9e-30 | 30.24% (291) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv1858 | modB | 2e-14 | 30.33% (211) | molbdenum-transport integral membrane protein ABC |
| M. leprae Br4923 | MLBr_01088 | - | 2e-09 | 23.21% (224) | putative ABC-transport protein, inner membrane component |
| M. abscessus ATCC 19977 | MAB_2434 | - | 2e-14 | 26.73% (217) | molybdenum ABC transporter permease |
| M. marinum M | MMAR_2732 | modB | 3e-15 | 28.38% (222) | molybdenum-transport integral membrane protein ABC |
| M. avium 104 | MAV_2861 | modB | 5e-16 | 29.09% (220) | molybdate ABC transporter, permease protein |
| M. thermoresistible (build 8) | TH_3819 | - | 1e-13 | 28.38% (222) | PUTATIVE NifC-like ABC-type porter |
| M. ulcerans Agy99 | MUL_3021 | modB | 2e-16 | 29.28% (222) | molybdenum-transport integral membrane protein ABC |
| M. vanbaalenii PYR-1 | Mvan_3389 | - | 2e-29 | 29.89% (271) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3136|M.gilvum_PYR-GCK MAGVATSSRQRSKIAPYLMVLPALAYLGIFFVVPFYSLARTSLSETAGSV
Mvan_3389|M.vanbaalenii_PYR-1 MAGVATSSRQRSKVAPYLMILPALAYLAIFFVVPFYSLARTSLSETAGSV
Mb1889|M.bovis_AF2122/97 --------MHPPTDLPRWVYLPAIAGIVFVAMPLVAIAIRVDWPRFWALI
Rv1858|M.tuberculosis_H37Rv --------MHPPTDLPRWVYLPAIAGIVFVAMPLVAIAIRVDWPRFWALI
MAV_2861|M.avium_104 ------------------MYLPAAAGTMFVVLPLLAIAVKVDWPHFWSLI
MMAR_2732|M.marinum_M --------MHGPTELPRWVYLPATAGMGLVILPLVAMAAKVDWPNFWSLI
MUL_3021|M.ulcerans_Agy99 --------MHGPPELPRWVYLPATAGMGLVILPLVAMAAKVDWPNFWSLI
TH_3819|M.thermoresistible__bu --------MNAST-LPRWIYLPAAVGATLLALPVFALAGRVDWPRFWSLI
MAB_2434|M.abscessus_ATCC_1997 --------MTPAPGLPRWVYVPAAIGAALIVLPLLALVVKADWGQFPALV
MSMEG_4763|M.smegmatis_MC2_155 ---MAAGQGLTDRARLAFLLTPPLAWLVVAYLGSLAVLLVSAFWQTNEFT
MLBr_01088|M.leprae_Br4923 ----MGIEKVKVTAGVRVGARRATIWVIIDTFVVGYALLPVLWILSLSLK
. . .
Mflv_3136|M.gilvum_PYR-GCK FMPTLTFAWDFGNYVDAFTLYREQIFRSFGYALVATLLCLLLAFPLAYVI
Mvan_3389|M.vanbaalenii_PYR-1 FMPTLTFAWDFGNYTEAFTLYREQIFRSFGYAFAATVLCLLLAFPLAYVI
Mb1889|M.bovis_AF2122/97 TTPSS----------------QTALLLSVKTAAASTVLCVLLGVPMALVL
Rv1858|M.tuberculosis_H37Rv TTPSS----------------QTALLLSVKTAAASTVLCVLLGVPMALVL
MAV_2861|M.avium_104 TSPSS----------------RTALLLSLRTAAASTALCVALGVPMALVL
MMAR_2732|M.marinum_M TSTSS----------------RTALVLSLKTASASTAICVLLGVPMALVL
MUL_3021|M.ulcerans_Agy99 TSTSS----------------RTALVLSLKTASASTAICVLLGVPTALVL
TH_3819|M.thermoresistible__bu CSESS----------------MTALALSLRTATASTVLCLVLGVPMALAL
MAB_2434|M.abscessus_ATCC_1997 TSPAS----------------QAALLLSVRTCLASTALCLLFGVPLAVVL
MSMEG_4763|M.smegmatis_MC2_155 GAVVRAFTLHNIERVLTDEVFRVATLRTLGVALTVTVICAALAVPIALYM
MLBr_01088|M.leprae_Br4923 PASTVK---------DGKLIPSLVTFDNYRGIFRSDLFSSALINSIGIGL
. :. : . . :
Mflv_3136|M.gilvum_PYR-GCK AFKAG-RFKNLILGLVILPFFVT---FLIRTIAWKTILADEGWLVSALGG
Mvan_3389|M.vanbaalenii_PYR-1 AFKAG-RFKNLILGLVILPFFVT---FLIRTIAWKTILADEGWVVSALGT
Mb1889|M.bovis_AF2122/97 ARSRG-RLVRSLRPLILLPLVLP---PVVGGIALLYAFGRLGLIGRYLEA
Rv1858|M.tuberculosis_H37Rv ARSRG-RLVRSLRPLILLPLVLP---PVVGGIALLYAFGRLGLIGRYLEA
MAV_2861|M.avium_104 ARGGT-RLVRSLRPLILLPLVLP---PVVGGIALLYAFGRLGLLGHYLEA
MMAR_2732|M.marinum_M ARSRA-RLMRLMRPLILLPLVLP---PVVGGIALLYAFGRLGLIGRYLEA
MUL_3021|M.ulcerans_Agy99 ARSRA-RLMRLMRPLILLPLVLP---PVVGGIALLYAFGRLGLIGRYLEA
TH_3819|M.thermoresistible__bu ARGGS-PILRWIRPLVLLPLVLP---PVVGGIALLYAFGRMGLIGGYLEA
MAB_2434|M.abscessus_ATCC_1997 ARGPG-RMTRLLRPMILLPLVLP---PVVGGIALLYAFGKLGLLGHYLDA
MSMEG_4763|M.smegmatis_MC2_155 AKIASPRARLILVVAVTTPLWAS---YLVKAYAWRMLLSPEGPLS---WA
MLBr_01088|M.leprae_Br4923 TTTVIAVMFGAMAAYAIARLAFPGKRLLVGTTLLVTMFPAISLVTPLFNI
: : : . :: : . :
Mflv_3136|M.gilvum_PYR-GCK IGLLPEEGRLLSTSWAVIGGLTYNWIIFMILPLYVSLEKIDPRLIEASKD
Mvan_3389|M.vanbaalenii_PYR-1 VGLLPEEGRLLSTSWAVIGGLTYNWIIFMILPLYVSLEKIDPRLIEASKD
Mb1889|M.bovis_AF2122/97 AGIS-----IAFSTAAVVLAQTFVSLPYLVISLEGAARTAGADYEVVAAT
Rv1858|M.tuberculosis_H37Rv AGIS-----IAFSTAAVVLAQTFVSLPYLVISLEGAARTAGADYEVVAAT
MAV_2861|M.avium_104 AGIS-----VAFSTTAVVLAQTFVSLPFLVISLEGAARTAGADFEVVAAT
MMAR_2732|M.marinum_M GGIT-----IAFSTTAVVLAQTFVSLPFLVVSLEGAARTAGADYEVVAAT
MUL_3021|M.ulcerans_Agy99 GGIT-----IAFSTTAVVLAQTFVSLPFLVVPLEGAARTAGADYEVVAAT
TH_3819|M.thermoresistible__bu VGIQ-----IAFSTTAVVLAQTFVSLPFLVLALEGAARTAGTGYEVVAAT
MAB_2434|M.abscessus_ATCC_1997 AGIR-----IAYSTSAVVLAQTFVSLPFLVISLQGALTTTGQRYDEVAAS
MSMEG_4763|M.smegmatis_MC2_155 TGFT-----PGYGIVATVATLTYLWLPYMVIPVYAAFERIPDALVDASSD
MLBr_01088|M.leprae_Br4923 ERRVG----LFDTWAGLILPYITFALPLAIYTLSAFFAEIPWDLEKAAKM
. : : : .: .:
Mflv_3136|M.gilvum_PYR-GCK LYSSTPRAFGKVILPLAMPGVLAGSMLVFIPAVGDFINADYLGSTQTTMI
Mvan_3389|M.vanbaalenii_PYR-1 LYSSTPRAFGKVILPLSMPGVLAGSMLVFIPAVGDFINADYLGSTQTTMI
Mb1889|M.bovis_AF2122/97 LGARPGTVWWRVTLPLLLPGVVSGSVLAFARSLGEFGATLTFAGSRQGVT
Rv1858|M.tuberculosis_H37Rv LGARPGTVWWRVTLPLLLPGVVSGSVLAFARSLGEFGATLTFAGSRQGVT
MAV_2861|M.avium_104 LGARPTTVWWRVTLPLLLPGVVSGAVLAFARSLGEFGATLTFAGSRQGVT
MMAR_2732|M.marinum_M LGARPSAVWWKVTLPLLMPGLVSGAVLAFARALGEFGATLTFAGSRQGVT
MUL_3021|M.ulcerans_Agy99 LGARPSAVWWKVTLPLLMPGLVSGAVLAFARSLGEFGATLTFAGSRQGVT
TH_3819|M.thermoresistible__bu LGATPTTVWWRVTLPLLAPGLVSGAVLAFARALGEFGATLTFAGSRQGVT
MAB_2434|M.abscessus_ATCC_1997 LGAPPTTVLRRVSLPLVMPALASGTVLAFARSLGEFGATLTFAGSRQGVT
MSMEG_4763|M.smegmatis_MC2_155 LGASDFATMRMVLAPLVFPGIAAGSIFTFSLSLGDYIAVTIVGG-KTQML
MLBr_01088|M.leprae_Br4923 DGATSGQAFRKVIVPLAAPGLVTAAILVFIFAWNDLLLALSLTSTKAAIT
: . * ** *.: :.:::.* : .: . . . : :
Mflv_3136|M.gilvum_PYR-GCK GNVIQKQFLVVKDYPAAAALSLGLMFLILVGVLLYTRALGTEDLV-----
Mvan_3389|M.vanbaalenii_PYR-1 GNVIQKQFLVVKDYPAAAALSLGLMLLILIGVLLYTRALGTEDLV-----
Mb1889|M.bovis_AF2122/97 R-TLPLEIYLQRVTDPDAAVALSLLLVVVAALVVLGVGAR--TPIGTDTR
Rv1858|M.tuberculosis_H37Rv R-TLPLEIYLQRVTDPDAAVALSLLLVVVAALVVLGVGAR--TPIGTDTR
MAV_2861|M.avium_104 R-TLPLEIYLQRVTDADAAVALSILLVAVAAVVVLGLGAR--RLTGTDAR
MMAR_2732|M.marinum_M R-TLPLEIYLQRVNDPDAAIALSLLLVAVAALVVLSLGAR--RLTGGQAG
MUL_3021|M.ulcerans_Agy99 R-TLPLEIYLQRVNDPDAAIALSLLLVAVAALVVLSLGAR--RLTGGQAG
TH_3819|M.thermoresistible__bu R-TLPLEIYLQREADPEAAVALSMVLIAVAAVVVIGLGAR--RLRAGGWS
MAB_2434|M.abscessus_ATCC_1997 R-TLPLEIYLQRESDPQAAVALSLLLVAVAAVVLAVVGARTRRPAGGGYE
MSMEG_4763|M.smegmatis_MC2_155 GNIIYGQLVTANNQPLAAALSIIPLAAIVAYLLAMRRTGALENV------
MLBr_01088|M.leprae_Br4923 APVAITNFTGSSQFEEPTGSIAAGAIVITAPIIVFVWIFQRRIVAGLTSG
:: :. ::
Mflv_3136|M.gilvum_PYR-GCK ----
Mvan_3389|M.vanbaalenii_PYR-1 ----
Mb1889|M.bovis_AF2122/97 ----
Rv1858|M.tuberculosis_H37Rv ----
MAV_2861|M.avium_104 ----
MMAR_2732|M.marinum_M ----
MUL_3021|M.ulcerans_Agy99 ----
TH_3819|M.thermoresistible__bu ----
MAB_2434|M.abscessus_ATCC_1997 ----
MSMEG_4763|M.smegmatis_MC2_155 ----
MLBr_01088|M.leprae_Br4923 AVKG