For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAGVATSSRQRSKVAPYLMILPALAYLAIFFVVPFYSLARTSLSETAGSVFMPTLTFAWDFGNYTEAFTL YREQIFRSFGYAFAATVLCLLLAFPLAYVIAFKAGRFKNLILGLVILPFFVTFLIRTIAWKTILADEGWV VSALGTVGLLPEEGRLLSTSWAVIGGLTYNWIIFMILPLYVSLEKIDPRLIEASKDLYSSTPRAFGKVIL PLSMPGVLAGSMLVFIPAVGDFINADYLGSTQTTMIGNVIQKQFLVVKDYPAAAALSLGLMLLILIGVLL YTRALGTEDLV
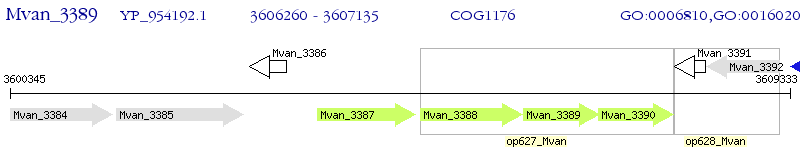
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3389 | - | - | 100% (291) | binding-protein-dependent transport systems inner membrane component |
| M. vanbaalenii PYR-1 | Mvan_2537 | - | 3e-12 | 24.88% (213) | NifC-like ABC-type porter |
| M. vanbaalenii PYR-1 | Mvan_6022 | - | 1e-10 | 25.00% (240) | binding-protein-dependent transport systems inner membrane |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1889 | modB | 1e-12 | 30.98% (255) | molbdenum-transport integral membrane protein ABC MODB |
| M. gilvum PYR-GCK | Mflv_3136 | - | 1e-157 | 95.19% (291) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv1858 | modB | 1e-12 | 30.98% (255) | molbdenum-transport integral membrane protein ABC |
| M. leprae Br4923 | MLBr_01088 | - | 8e-08 | 26.25% (259) | putative ABC-transport protein, inner membrane component |
| M. abscessus ATCC 19977 | MAB_2434 | - | 3e-13 | 25.45% (224) | molybdenum ABC transporter permease |
| M. marinum M | MMAR_2732 | modB | 9e-13 | 30.49% (246) | molybdenum-transport integral membrane protein ABC |
| M. avium 104 | MAV_2861 | modB | 4e-10 | 26.10% (249) | molybdate ABC transporter, permease protein |
| M. smegmatis MC2 155 | MSMEG_3279 | - | 1e-135 | 86.45% (273) | polyamine ABC transporter permease protein |
| M. thermoresistible (build 8) | TH_3819 | - | 3e-10 | 24.50% (200) | PUTATIVE NifC-like ABC-type porter |
| M. ulcerans Agy99 | MUL_3021 | modB | 8e-13 | 27.20% (239) | molybdenum-transport integral membrane protein ABC |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_3389|M.vanbaalenii_PYR-1 --------MAGVATSSRQRSKVAPYLMILPALAYLAIFFVVPFYSLARTS
Mflv_3136|M.gilvum_PYR-GCK --------MAGVATSSRQRSKIAPYLMVLPALAYLGIFFVVPFYSLARTS
MSMEG_3279|M.smegmatis_MC2_155 --------------------------MILPALVYLGIFFVVPFFTLARTS
Mb1889|M.bovis_AF2122/97 ----------------------MHPPTDLPRWVYLPAIAGIVFVAMPLVA
Rv1858|M.tuberculosis_H37Rv ----------------------MHPPTDLPRWVYLPAIAGIVFVAMPLVA
MAV_2861|M.avium_104 --------------------------------MYLPAAAGTMFVVLPLLA
MMAR_2732|M.marinum_M ----------------------MHGPTELPRWVYLPATAGMGLVILPLVA
MUL_3021|M.ulcerans_Agy99 ----------------------MHGPPELPRWVYLPATAGMGLVILPLVA
TH_3819|M.thermoresistible__bu ----------------------MNAST-LPRWIYLPAAVGATLLALPVFA
MAB_2434|M.abscessus_ATCC_1997 ----------------------MTPAPGLPRWVYVPAAIGAALIVLPLLA
MLBr_01088|M.leprae_Br4923 MGIEKVKVTAGVRVGARRATIWVIIDTFVVGYALLPVLWILSLSLKPAST
: : . :
Mvan_3389|M.vanbaalenii_PYR-1 LSETAGSVFMPTLTFAWDFGNYTEAFTLYREQIFRSFGYAFAATVLCLLL
Mflv_3136|M.gilvum_PYR-GCK LSETAGSVFMPTLTFAWDFGNYVDAFTLYREQIFRSFGYALVATLLCLLL
MSMEG_3279|M.smegmatis_MC2_155 LSSSGGSIYLPTLTFDWNFGNYLHAFSAYQDQILRSFGYAFVATVLCVIL
Mb1889|M.bovis_AF2122/97 IAIR----------VDWPRFWALITTPSSQTALLLSVKTAAASTVLCVLL
Rv1858|M.tuberculosis_H37Rv IAIR----------VDWPRFWALITTPSSQTALLLSVKTAAASTVLCVLL
MAV_2861|M.avium_104 IAVK----------VDWPHFWSLITSPSSRTALLLSLRTAAASTALCVAL
MMAR_2732|M.marinum_M MAAK----------VDWPNFWSLITSTSSRTALVLSLKTASASTAICVLL
MUL_3021|M.ulcerans_Agy99 MAAK----------VDWPNFWSLITSTSSRTALVLSLKTASASTAICVLL
TH_3819|M.thermoresistible__bu LAGR----------VDWPRFWSLICSESSMTALALSLRTATASTVLCLVL
MAB_2434|M.abscessus_ATCC_1997 LVVK----------ADWGQFPALVTSPASQAALLLSVRTCLASTALCLLF
MLBr_01088|M.leprae_Br4923 VKDGK----LIPSLVTFDNYRGIFRSDLFSSALINSIGIGLTTTVIAVMF
: : : *. .:* :.: :
Mvan_3389|M.vanbaalenii_PYR-1 AFPLAYVIAFKAGRFKNLILGLVILPFFVTFLIRTIAWKTILADEGWVVS
Mflv_3136|M.gilvum_PYR-GCK AFPLAYVIAFKAGRFKNLILGLVILPFFVTFLIRTIAWKTILADEGWLVS
MSMEG_3279|M.smegmatis_MC2_155 AFPLAYVIAFKAGRFKNLILGLVILPFFVTFLIRTIAWKTILADDGWVVS
Mb1889|M.bovis_AF2122/97 GVPMALVLARSRGRLVRSLRPLILLPLVLPPVVGGIALLYAFGRLGLIGR
Rv1858|M.tuberculosis_H37Rv GVPMALVLARSRGRLVRSLRPLILLPLVLPPVVGGIALLYAFGRLGLIGR
MAV_2861|M.avium_104 GVPMALVLARGGTRLVRSLRPLILLPLVLPPVVGGIALLYAFGRLGLLGH
MMAR_2732|M.marinum_M GVPMALVLARSRARLMRLMRPLILLPLVLPPVVGGIALLYAFGRLGLIGR
MUL_3021|M.ulcerans_Agy99 GVPTALVLARSRARLMRLMRPLILLPLVLPPVVGGIALLYAFGRLGLIGR
TH_3819|M.thermoresistible__bu GVPMALALARGGSPILRWIRPLVLLPLVLPPVVGGIALLYAFGRMGLIGG
MAB_2434|M.abscessus_ATCC_1997 GVPLAVVLARGPGRMTRLLRPMILLPLVLPPVVGGIALLYAFGKLGLLGH
MLBr_01088|M.leprae_Br4923 GAMAAYAIARLAFPGKRLLVGTTLLVTMFPAISLVTPLFNIERRVGLFDT
. * .:* . : :* ... : . * .
Mvan_3389|M.vanbaalenii_PYR-1 ALGTVGLLPEEGRLLSTSWAVIGGLTYNWIIFMILPLYVSLEKIDPRLIE
Mflv_3136|M.gilvum_PYR-GCK ALGGIGLLPEEGRLLSTSWAVIGGLTYNWIIFMILPLYVSLEKIDPRLIE
MSMEG_3279|M.smegmatis_MC2_155 ALGAIGLLPDEGRLLSTPWAVIGGLTYNWIIFMILPLYVSLEKIDPRLIE
Mb1889|M.bovis_AF2122/97 YLEAAGIS-----IAFSTAAVVLAQTFVSLPYLVISLEGAARTAGADYEV
Rv1858|M.tuberculosis_H37Rv YLEAAGIS-----IAFSTAAVVLAQTFVSLPYLVISLEGAARTAGADYEV
MAV_2861|M.avium_104 YLEAAGIS-----VAFSTTAVVLAQTFVSLPFLVISLEGAARTAGADFEV
MMAR_2732|M.marinum_M YLEAGGIT-----IAFSTTAVVLAQTFVSLPFLVVSLEGAARTAGADYEV
MUL_3021|M.ulcerans_Agy99 YLEAGGIT-----IAFSTTAVVLAQTFVSLPFLVVPLEGAARTAGADYEV
TH_3819|M.thermoresistible__bu YLEAVGIQ-----IAFSTTAVVLAQTFVSLPFLVLALEGAARTAGTGYEV
MAB_2434|M.abscessus_ATCC_1997 YLDAAGIR-----IAYSTSAVVLAQTFVSLPFLVISLQGALTTTGQRYDE
MLBr_01088|M.leprae_Br4923 WAG-----------------LILPYITFALPLAIYTLSAFFAEIPWDLEK
:: : : .*
Mvan_3389|M.vanbaalenii_PYR-1 ASKDLYSSTPRAFGKVILPLSMPGVLAGSMLVFIPAVGDF-INADYLGST
Mflv_3136|M.gilvum_PYR-GCK ASKDLYSSTPRAFGKVILPLAMPGVLAGSMLVFIPAVGDF-INADYLGST
MSMEG_3279|M.smegmatis_MC2_155 ASKDLYSSNTRSFTKVILPLSMPGVLAGSMLVFIPAVGDF-INADYLGST
Mb1889|M.bovis_AF2122/97 VAATLGARPGTVWWRVTLPLLLPGVVSGSVLAFARSLGEFGATLTFAGSR
Rv1858|M.tuberculosis_H37Rv VAATLGARPGTVWWRVTLPLLLPGVVSGSVLAFARSLGEFGATLTFAGSR
MAV_2861|M.avium_104 VAATLGARPTTVWWRVTLPLLLPGVVSGAVLAFARSLGEFGATLTFAGSR
MMAR_2732|M.marinum_M VAATLGARPSAVWWKVTLPLLMPGLVSGAVLAFARALGEFGATLTFAGSR
MUL_3021|M.ulcerans_Agy99 VAATLGARPSAVWWKVTLPLLMPGLVSGAVLAFARSLGEFGATLTFAGSR
TH_3819|M.thermoresistible__bu VAATLGATPTTVWWRVTLPLLAPGLVSGAVLAFARALGEFGATLTFAGSR
MAB_2434|M.abscessus_ATCC_1997 VAASLGAPPTTVLRRVSLPLVMPALASGTVLAFARSLGEFGATLTFAGSR
MLBr_01088|M.leprae_Br4923 AAKMDGATSGQAFRKVIVPLAAPGLVTAAILVFIFAWNDLLLALSLTSTK
.: : :* :** *.: :.::*.* : .:: .:
Mvan_3389|M.vanbaalenii_PYR-1 QTTMIGNVIQKQFL--VVKDYPAAAALSLGLMLLILIGVLLYTRA---LG
Mflv_3136|M.gilvum_PYR-GCK QTTMIGNVIQKQFL--VVKDYPAAAALSLGLMFLILVGVLLYTRA---LG
MSMEG_3279|M.smegmatis_MC2_155 QTTMIGNVIQKQFL--VVKDYPAAAALSMVLMAIILVGVLMYTRA---LG
Mb1889|M.bovis_AF2122/97 QGVTRTLPLEIYLQ--RVTDPDAAVALSLLLVVVAALVVLGVGAR--TPI
Rv1858|M.tuberculosis_H37Rv QGVTRTLPLEIYLQ--RVTDPDAAVALSLLLVVVAALVVLGVGAR--TPI
MAV_2861|M.avium_104 QGVTRTLPLEIYLQ--RVTDADAAVALSILLVAVAAVVVLGLGAR--RLT
MMAR_2732|M.marinum_M QGVTRTLPLEIYLQ--RVNDPDAAIALSLLLVAVAALVVLSLGAR--RLT
MUL_3021|M.ulcerans_Agy99 QGVTRTLPLEIYLQ--RVNDPDAAIALSLLLVAVAALVVLSLGAR--RLT
TH_3819|M.thermoresistible__bu QGVTRTLPLEIYLQ--READPEAAVALSMVLIAVAAVVVIGLGAR--RLR
MAB_2434|M.abscessus_ATCC_1997 QGVTRTLPLEIYLQ--RESDPQAAVALSLLLVAVAAVVLAVVGARTRRPA
MLBr_01088|M.leprae_Br4923 AAITAPVAITNFTGSSQFEEPTGSIAAGAIVITAPIIVFVWIFQRRIVAG
: : .: * . :: : .
Mvan_3389|M.vanbaalenii_PYR-1 TEDLV---
Mflv_3136|M.gilvum_PYR-GCK TEDLV---
MSMEG_3279|M.smegmatis_MC2_155 TEDLV---
Mb1889|M.bovis_AF2122/97 GTDTR---
Rv1858|M.tuberculosis_H37Rv GTDTR---
MAV_2861|M.avium_104 GTDAR---
MMAR_2732|M.marinum_M GGQAG---
MUL_3021|M.ulcerans_Agy99 GGQAG---
TH_3819|M.thermoresistible__bu AGGWS---
MAB_2434|M.abscessus_ATCC_1997 GGGYE---
MLBr_01088|M.leprae_Br4923 LTSGAVKG