For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPPTPRLQAGDTAPAFSLPDADGNTVSLSDYRGRKVIVYFYPAASTPGCTKQACDFRDNLAELNDAGLDV VGISPDKPEKLAKFRDKEGLTFPLLSDPDRSVLTAWGAYGEKTMYGKTVQGVIRSTFLVDEDGKIAVAQY NVRATGHVAKLRRDLSV
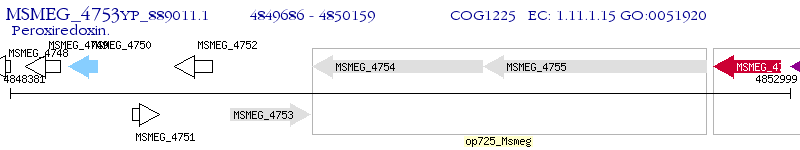
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4753 | - | - | 100% (157) | antioxidant, AhpC/TSA family protein |
| M. smegmatis MC2 155 | MSMEG_3216 | - | 7e-23 | 41.96% (143) | peroxiredoxin Q |
| M. smegmatis MC2 155 | MSMEG_4320 | - | 7e-12 | 33.81% (139) | alkyl hydroperoxide reductase/ Thiol specific antioxidant/ Mal |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2550 | bcp | 1e-74 | 85.06% (154) | bacterioferritin comigratory protein BCP |
| M. gilvum PYR-GCK | Mflv_2535 | - | 3e-79 | 87.90% (157) | alkyl hydroperoxide reductase/ Thiol specific antioxidant/ Mal |
| M. tuberculosis H37Rv | Rv2521 | bcp | 7e-75 | 85.71% (154) | bacterioferritin comigratory protein BCP |
| M. leprae Br4923 | MLBr_00424 | bcp | 7e-67 | 78.57% (154) | putative antioxidant protein |
| M. abscessus ATCC 19977 | MAB_1515c | - | 4e-69 | 76.43% (157) | Alkyl hydroperoxide reductase/ Thiol specific antioxidant/ Mal |
| M. marinum M | MMAR_3956 | bcp | 8e-75 | 86.36% (154) | bacterioferritin comigratory protein Bcp |
| M. avium 104 | MAV_1653 | - | 8e-75 | 87.66% (154) | alkyl hydroperoxide reductase/ Thiol specific antioxidant/ Mal |
| M. thermoresistible (build 8) | TH_1100 | bcp | 8e-79 | 89.61% (154) | PROBABLE BACTERIOFERRITIN COMIGRATORY PROTEIN BCP |
| M. ulcerans Agy99 | MUL_3811 | bcp | 5e-75 | 86.36% (154) | bacterioferritin comigratory protein Bcp |
| M. vanbaalenii PYR-1 | Mvan_4120 | - | 1e-79 | 88.54% (157) | redoxin domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4753|M.smegmatis_MC2_155 MPPTPRLQAGDTAPAFSLPDADGNTVSLSDYRGRKVIVYFYPAASTPGCT
TH_1100|M.thermoresistible__bu VAQTQRLEVGDPAPPFSLPDADGNTVSLADYRGRKLIVYFYPAASTPGCT
Mvan_4120|M.vanbaalenii_PYR-1 MPQTPRLEVGDTAPAFSLPDADGNTVKLADYKGRKVIVYFYPAASTPGCT
Mflv_2535|M.gilvum_PYR-GCK MPQTPRLEVGDKAPTFSLPDADGNTVSLSDYKGRKVIVYFYPAASTPGCT
MAB_1515c|M.abscessus_ATCC_199 MAETKRLEVGDKAPTFTLLDADGKKVSLSSYKGRKVIIYFYPAAMTPGCT
Mb2550|M.bovis_AF2122/97 MTKTTRLTPGDKAPAFTLPDADGNNVSLADYRGRRVIVYFYPAASTPGCT
Rv2521|M.tuberculosis_H37Rv MTKTTRLTPGDKAPAFTLPDADGNNVSLADYRGRRVIVYFYPAASTPGCT
MLBr_00424|M.leprae_Br4923 MTETTRLAPGDQAPAFSLPDADGRIVSLGDYRGQRVTVYFYPAALTPGCT
MMAR_3956|M.marinum_M MSETTRLAPGDKAPAFSLPDANGNKVSLSDYKGRQVVVYFYPAASTPGCT
MUL_3811|M.ulcerans_Agy99 MSETTRLAPGDKAPAFSLPDANGNKVSLSDYKGRQVVVYFYPAASTPGCT
MAV_1653|M.avium_104 MTETTRLAPGDKAPAFSLPDADGNTVKLSDFKGRKVIVYFYPAASTPGCT
:. * ** ** **.*:* **:*. *.*..::*::: :****** *****
MSMEG_4753|M.smegmatis_MC2_155 KQACDFRDNLAELNDAGLDVVGISPDKPEKLAKFRDKEGLTFPLLSDPDR
TH_1100|M.thermoresistible__bu KQACDFRDNLAELNEAGLDVVGISPDKPEKLAKFRDAQGLTFPLLSDPDR
Mvan_4120|M.vanbaalenii_PYR-1 KEACDFRDSLGELNEAGLDVVGISPDKPEKLAKFRDAENLTFPLLSDPDR
Mflv_2535|M.gilvum_PYR-GCK KQACDFRDSLAELNDAGLDIIGISPDKPEKLAKFRDAEKLTFPLLSDPEK
MAB_1515c|M.abscessus_ATCC_199 KQACDFRDSLAELNEAGLDVIGISPDKPEKLAKFRDRDGVNFPLLSDPDK
Mb2550|M.bovis_AF2122/97 KQACDFRDNLGDFTTAGLNVVGISPDKPEKLATFRDAQGLTFPLLSDPDR
Rv2521|M.tuberculosis_H37Rv KQACDFRDNLGDFTTAGLNVVGISPDKPEKLATFRDAQGLTFPLLSDPDR
MLBr_00424|M.leprae_Br4923 KQACDFRDNLHDLNDAGLDVVGISPDPLTKLAEFRDAEALTFPLLSDHDC
MMAR_3956|M.marinum_M KQACDFRDNLSELNEAGLDVVGISPDKPEKLAKFRDAEGLTFPLLSDPDR
MUL_3811|M.ulcerans_Agy99 KQACDFRDNLSELNEAGLDVVGISPDKPEKLAKFRDAEGLTFPLLSDPDR
MAV_1653|M.avium_104 KQACDFRDSLAELNGAGLDVVGISPDKPEKLAKFRDAEKLTFPLLSDPDR
*:******.* ::. ***:::***** *** *** : :.****** :
MSMEG_4753|M.smegmatis_MC2_155 SVLTAWGAYGEKTMYGKTVQGVIRSTFLVDEDGKIAVAQYNVRATGHVAK
TH_1100|M.thermoresistible__bu EVLTAWGAFGEKKMYGKTVQGVIRSTFVVDEDGKIALAQYNVRATGHVAK
Mvan_4120|M.vanbaalenii_PYR-1 KVLEAWGAYGEKKMYGKTVQGVIRSTFVVDEKGKIAVAQYNIRATGHVAK
Mflv_2535|M.gilvum_PYR-GCK KVLEAWGAFGEKTMYGKTVQGVIRSTFVVDENGKIEVAQYNVRATGHVAK
MAB_1515c|M.abscessus_ATCC_199 TTLTAYGAFGEKKLYGKIVEGVIRSTFVVDEKGNIEVAQYNVKATGHVAK
Mb2550|M.bovis_AF2122/97 EVLTAWGAYGEKQMYGKTVQGMIRSTFVVDEDGKIVVAQYNVKATGHVAK
Rv2521|M.tuberculosis_H37Rv EVLTAWGAYGEKQMYGKTVQGVIRSTFVVDEDGKIVVAQYNVKATGHVAK
MLBr_00424|M.leprae_Br4923 KVLSAWGAYGKKQIYGKTVLGVIRSTFVVDEKGKIVLAQYNVKATGHVAK
MMAR_3956|M.marinum_M KVLAAYGAYGEKMMYGKTVTGVIRSTFVVDENGKIAVAQYNVKATGHVAK
MUL_3811|M.ulcerans_Agy99 KVLAAYGAYGEKMMYGKTVTGVIRSTFVVDENGKIAVAQYNVKATGHVAK
MAV_1653|M.avium_104 KVLTAYGAYGEKQMYGKTVTGVIRSTFVVDEKGRIAVAQYNVKATGHVAK
.* *:**:*:* :*** * *:*****:***.*.* :****::*******
MSMEG_4753|M.smegmatis_MC2_155 LRRDLSV----
TH_1100|M.thermoresistible__bu LRRDLGV----
Mvan_4120|M.vanbaalenii_PYR-1 LRRDLSV----
Mflv_2535|M.gilvum_PYR-GCK LRRDLSV----
MAB_1515c|M.abscessus_ATCC_199 LRRDISV----
Mb2550|M.bovis_AF2122/97 LRRDLSV----
Rv2521|M.tuberculosis_H37Rv LRRDLSV----
MLBr_00424|M.leprae_Br4923 LRRDLSLLECC
MMAR_3956|M.marinum_M LRRDLQV----
MUL_3811|M.ulcerans_Agy99 LRRDLQV----
MAV_1653|M.avium_104 LRRDLSV----
****: :