For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSIGTGDTVAEFELPDQTGQKRSLTRLLGDGPVVLFFYPAAMTPGCTKEACHFRDLAQEFAAVGANPVGI STDPPAKQARFAETHRFAFPLLSDADGTVAARFGVKRGLLGKLMPVKRTTFVIDTDSTVLAVISSEINMD THADKALEVLRSR*
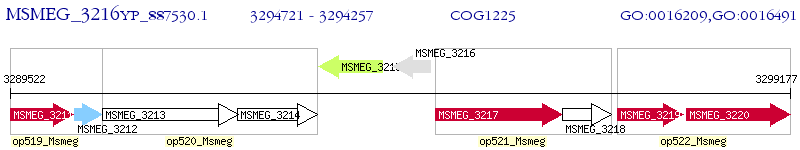
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3216 | - | - | 100% (154) | peroxiredoxin Q |
| M. smegmatis MC2 155 | MSMEG_4753 | - | 7e-23 | 41.96% (143) | antioxidant, AhpC/TSA family protein |
| M. smegmatis MC2 155 | MSMEG_4320 | - | 2e-05 | 28.03% (132) | alkyl hydroperoxide reductase/ Thiol specific antioxidant/ Mal |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1634c | bcpB | 4e-63 | 79.73% (148) | peroxidoxin bcpB |
| M. gilvum PYR-GCK | Mflv_3603 | - | 5e-64 | 78.15% (151) | alkyl hydroperoxide reductase/ Thiol specific antioxidant/ Mal |
| M. tuberculosis H37Rv | Rv1608c | bcpB | 4e-63 | 79.73% (148) | peroxidoxin BcpB |
| M. leprae Br4923 | MLBr_00424 | bcp | 3e-22 | 42.11% (133) | putative antioxidant protein |
| M. abscessus ATCC 19977 | MAB_2662 | - | 7e-57 | 70.27% (148) | peroxidoxin BcpB |
| M. marinum M | MMAR_2410 | bcpB | 5e-63 | 78.38% (148) | peroxidoxin BcpB |
| M. avium 104 | MAV_3178 | - | 2e-63 | 79.73% (148) | antioxidant, AhpC/TSA family protein |
| M. thermoresistible (build 8) | TH_2740 | - | 3e-55 | 68.92% (148) | PUTATIVE alkyl hydroperoxide reductase/ Thiol specific |
| M. ulcerans Agy99 | MUL_1588 | bcpB | 3e-63 | 78.38% (148) | peroxidoxin BcpB |
| M. vanbaalenii PYR-1 | Mvan_2813 | - | 4e-62 | 79.31% (145) | redoxin domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1634c|M.bovis_AF2122/97 ------MKTGDTVADFELPDQTGTPRRLSVLLSDGPVVLFFYPAAMTPGC
Rv1608c|M.tuberculosis_H37Rv ------MKTGDTVADFELPDQTGTPRRLSVLLSDGPVVLFFYPAAMTPGC
MMAR_2410|M.marinum_M ------MKIGDTVADFELPDQTGKQRKLSALLSDGPVVLFFYPAAMTPGC
MUL_1588|M.ulcerans_Agy99 ------MKIGDTVADFELPDQTGKQRKLSALLSDGPVVLFFYPAAMTPGC
MAV_3178|M.avium_104 ------MKPGDTVADFELPDQTGTPRKLSDLLAAGPVVLFFYPAAMTPGC
Mflv_3603|M.gilvum_PYR-GCK ---MEIVNRGDRVTEFELPDQTGTTRTLTGLLADGPVVLFFYPAAMTPGC
Mvan_2813|M.vanbaalenii_PYR-1 ------------MAEFELPDQTGTTRTLTGLLADGPVVLFFYPAAMTPGC
MSMEG_3216|M.smegmatis_MC2_155 ---MTSIGTGDTVAEFELPDQTGQKRSLTRLLGDGPVVLFFYPAAMTPGC
MAB_2662|M.abscessus_ATCC_1997 ------MKPGDRVADFELPDQTGTSRSLSALLADGPVVLFFYPAANTPGC
TH_2740|M.thermoresistible__bu ------MKPGEPAPEFELPDQTGAVQSLSTLLAAGPVALFFYPAALTPGC
MLBr_00424|M.leprae_Br4923 MTETTRLAPGDQAPAFSLPDADGRIVSLGDYRGQR-VTVYFYPAALTPGC
. *.*** * * . *.::***** ****
Mb1634c|M.bovis_AF2122/97 TKEACHFRDLAKEFAEVRASRVGISTDPVRKQAKFAEVRRFDYPLLSDAQ
Rv1608c|M.tuberculosis_H37Rv TKEACHFRDLAKEFAEVRASRVGISTDPVRKQAKFAEVRRFDYPLLSDAQ
MMAR_2410|M.marinum_M TKEACHFRDLAAEFTAVGASRVGISADSVQKQAKFADMQHFDYPLLSDAE
MUL_1588|M.ulcerans_Agy99 TKEACHFRDLAAEFTAVGASRVGISADSVQKQAKFADMQHFDYPLLSDAE
MAV_3178|M.avium_104 TKEACHFRDLAAEFAAVGANRVGISADPVDKQAKFAELQKFDYPLLSDTD
Mflv_3603|M.gilvum_PYR-GCK TKEACHFRDLAAEFAAVGATRVGISADPVDKQAQFADKQNFDYPLLSDTE
Mvan_2813|M.vanbaalenii_PYR-1 TKEACHFRDLAAEFAAVGATRVGISADPVDKQAKFADQQKFDYPLLSDTE
MSMEG_3216|M.smegmatis_MC2_155 TKEACHFRDLAQEFAAVGANPVGISTDPPAKQARFAETHRFAFPLLSDAD
MAB_2662|M.abscessus_ATCC_1997 TAEACHFRDLASEFKEVGASRVGISVDSVEKQADFADKRKFDYPLLSDTG
TH_2740|M.thermoresistible__bu TKQARHFRDLAADFADAGATPVGISVDPVAKQAKFADIENFEFPLLSDRD
MLBr_00424|M.leprae_Br4923 TKQACDFRDNLHDLNDAGLDVVGISPDPLTKLAEFRDAEALTFPLLSDHD
* :* .*** :: . **** *. * * * : . : :*****
Mb1634c|M.bovis_AF2122/97 GTVAAQFGV--KRGLLGK-LMPVKRTTFVIDTDRKVLDVISSEFSMDAHA
Rv1608c|M.tuberculosis_H37Rv GTVAAQFGV--KRGLLGK-LMPVKRTTFVIDTDRKVLDVISSEFSMDAHA
MMAR_2410|M.marinum_M GTVAGQFGV--KRGLLGK-LMPVKRTTFVIDTDRKVLNVISSEFSMDTHA
MUL_1588|M.ulcerans_Agy99 GTVAGQFGV--KRGLLGK-LMPVKRTTFVIDTDRKVLNVISSEFSMDTHA
MAV_3178|M.avium_104 GAVAAQFGV--KRGLLSK-LLPVKRTTFVIDTDRTVLDVISSEFNMDTHA
Mflv_3603|M.gilvum_PYR-GCK GTVAAQFGV--KRGLLGK-FMPVKRTTFVIDTDRTVLEVIASEFSMDTHA
Mvan_2813|M.vanbaalenii_PYR-1 GVVATQFGV--KRGLLGK-FMPVKRTTFVIDTDRTVLEVIASEFSMDTHA
MSMEG_3216|M.smegmatis_MC2_155 GTVAARFGV--KRGLLGK-LMPVKRTTFVIDTDSTVLAVISSEINMDTHA
MAB_2662|M.abscessus_ATCC_1997 GKISSAFGV--KRGLLGK-LAPVKRTTFIIDTDKTILEVFASELNMNAHA
TH_2740|M.thermoresistible__bu GAVARRYGV--KRSILGK-LLPVRRTTFVIDTDRTVLEVIASELNMAVHA
MLBr_00424|M.leprae_Br4923 CKVLSAWGAYGKKQIYGKTVLGVIRSTFVVD-EKGKIVLAQYNVKATGHV
: :*. *: : .* . * *:**::* : : : :.. *.
Mb1634c|M.bovis_AF2122/97 DKALATLRAIRSG
Rv1608c|M.tuberculosis_H37Rv DKALATLRAIRSG
MMAR_2410|M.marinum_M DKALETLRARA--
MUL_1588|M.ulcerans_Agy99 DKALETLRARA--
MAV_3178|M.avium_104 DKALATLRARA--
Mflv_3603|M.gilvum_PYR-GCK DKALEVLRAR---
Mvan_2813|M.vanbaalenii_PYR-1 DKALEVLRGRQSA
MSMEG_3216|M.smegmatis_MC2_155 DKALEVLRSR---
MAB_2662|M.abscessus_ATCC_1997 DKALEFLRARK--
TH_2740|M.thermoresistible__bu EKALAALRDRQSA
MLBr_00424|M.leprae_Br4923 AKLRRDLSLLECC
* *