For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLNSAATPDNRLSFIDQAGFDMLRATGRHQLMQVLWIYEHAIDMDGLRQFHRNFGYGLAGRRIERSRLPF GRHRWVSCPGPQTELAVATTARPRADVSDWADEQVHLPVDPEHGPAWHMSVLPMTDGSTAVSLVASHHIG DGIGGVLTLMEAVAGTKRQFGFPPPRAQTARRAVAADLRQTARDLPELGRTAGKAVRLLRRRGAELLRSA RAENESAPRTLSTEPVSVPAVSVFVDLDAWDRCATGLGGNSYSLLAGFGARLGERMNRANARSGTVTLSI ALNERTSLDDSRALAMTFASASIDPRQCTSDLTGVRATIREAIETARTTTDETLELLPLIPLVPKRALQG LGGLFFGSPDDLPVYCSNLGDVDSAVGRADGTDAEYVVMRGVDQNVTRRDIEAMGGQLVLVSVRVNGKIS IGIVAYQLDAQNSKTRLRELAALTLKDFGLAGVIE
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
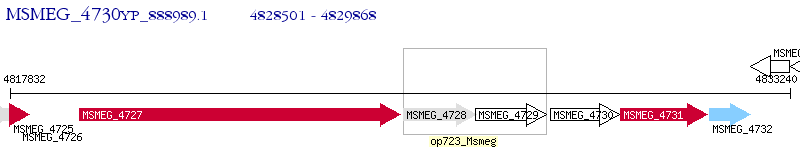
| M. smegmatis MC2 155 | MSMEG_4730 | - | - | 100% (455) | hypothetical protein MSMEG_4730 |
| M. smegmatis MC2 155 | MSMEG_4729 | - | e-113 | 48.21% (446) | hypothetical protein MSMEG_4729 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4701 | - | 1e-102 | 44.30% (456) | hypothetical protein MAB_4701 |
| M. marinum M | MMAR_2354 | - | 2e-98 | 42.35% (451) | hypothetical protein MMAR_2354 |
| M. avium 104 | MAV_4881 | - | 8e-90 | 43.02% (444) | hypothetical protein MAV_4881 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1530 | - | 1e-97 | 42.13% (451) | hypothetical protein MUL_1530 |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2354|M.marinum_M ----------MLNESRTRSSSAMTDNRIAFLDQAAFLRLRATGFETVGQT
MUL_1530|M.ulcerans_Agy99 ----------MLNESRTRSSAAMTDNRIAFLDQAAFLRLRATGFETVGQT
MAV_4881|M.avium_104 -----------------------MSNVLDLADQTLFLGERATGATSLLQC
MSMEG_4730|M.smegmatis_MC2_155 ----------------MLNSAATPDNRLSFIDQAGFDMLRATGRHQLMQV
MAB_4701|M.abscessus_ATCC_1997 MPGQIRHMRKSLSRRRTSKGAAAMDNRLALADEALLAEHHAAGMNVIIQV
.* : : *:: : :*:* : *
MMAR_2354|M.marinum_M LWIYDREADMDGLRRFNENLGYGLLGRRIERSPLPFGRPRWVAYHEA-PD
MUL_1530|M.ulcerans_Agy99 LWIYDREADMDGLRRSNENLGYGLLGRRIERSPLPFGRPRWVAYHEA-PD
MAV_4881|M.avium_104 VWVYDRAIDLAGLRRFHRHLGAGRLARRIECSPLPFGRHRWVSTGAQ-GA
MSMEG_4730|M.smegmatis_MC2_155 LWIYEHAIDMDGLRQFHRNFGYGLAGRRIERSRLPFGRHRWVSCPGPQTE
MAB_4701|M.abscessus_ATCC_1997 TWLYSHAIDLDALQRFRHNLGRGLLGRRIEAPALPIARHRWVIDHGP-AD
*:*.: *: .*:: ..::* * .**** . **:.* ***
MMAR_2354|M.marinum_M IDVAATPRPRSEITAWLDERAQIPVDPEYGPCWHLGVLPLQDGGTAVSVV
MUL_1530|M.ulcerans_Agy99 IDVAATPRPRSEITAWLDERAQIPVDPEYGPCWHLGVLPLQDGGTAVSVV
MAV_4881|M.avium_104 LEVVAVPRPRDAVDAWLTEQAGRRPDAEHGPGWHLAVLPLTDGGAVVSLV
MSMEG_4730|M.smegmatis_MC2_155 LAVATTARPRADVSDWADEQVHLPVDPEHGPAWHMSVLPMTDGSTAVSLV
MAB_4701|M.abscessus_ATCC_1997 IDIEERARPRAEFSEWVDERSQLHIDPESGPGWHIGVLPLQDGTTAISLV
: : .*** . * *: *.* ** **:.***: ** :.:*:*
MMAR_2354|M.marinum_M ITHSVVDGVAGGLAVFEAINGITRDFGYPPPGSRGRMEGLLVDLWDALRG
MUL_1530|M.ulcerans_Agy99 ITHSVVDGVAGGLAVFEAINGITRDFGYPPPGSRGRMEGLLVDLWDALRG
MAV_4881|M.avium_104 TTHCLTDGVGLCEALADAARGRADTVRWPAAAARPRWSALAEDARQTARD
MSMEG_4730|M.smegmatis_MC2_155 ASHHIGDGIGGVLTLMEAVAGTKRQFGFPPPRAQTARRAVAADLRQTARD
MAB_4701|M.abscessus_ATCC_1997 LSHNLIDGLGLALVIVEAVMGRTRDLGYPLPLSRRRLRALGQDVRQTARD
:* : **:. .: :* * . :* . :: .: * :: *.
MMAR_2354|M.marinum_M LPEVFRALLALILIVLR-----RDTGAAGQTGKPPTVSDGADEPIVTPSV
MUL_1530|M.ulcerans_Agy99 LPEVFRALLALILIVLR-----RDTGAAGQTGKPPTVSDGADEPIVTPSV
MAV_4881|M.avium_104 TGDVGRAVRAAARMLRQ-----GRGGAALARGR-ATRRRGTQDRVTVPAQ
MSMEG_4730|M.smegmatis_MC2_155 LPELGRTAGKAVRLLRR--RGAELLRSARAENESAPRTLST-EPVSVPAV
MAB_4701|M.abscessus_ATCC_1997 VPQATRALVAAVRLGRKQARDKNLPGLAPVAVRPAGSTAPSDEVVLVPAI
: *: : : * . . : : : .*:
MMAR_2354|M.marinum_M TAFCDLETWDSCALDLSGSSGSLFVSFATRLAQLMGRVRATDGAVTIIMP
MUL_1530|M.ulcerans_Agy99 TAFCDLETWDSCALDLSGSSGSLFVSFATRLAQLMGRVRVTDGAVTIIMP
MAV_4881|M.avium_104 TIFVDADEWDARARALGGTSNALLAGLAARLAQRVGRV-ARDGSVTVGMP
MSMEG_4730|M.smegmatis_MC2_155 SVFVDLDAWDRCATGLGGNSYSLLAGFGARLGERMNRANARSGTVTLSIA
MAB_4701|M.abscessus_ATCC_1997 TIYVGLDAWDARAEALGGASHTLAAAFAARFGERIGRRRAHDGAITLQIP
: : . : ** * *.* * :* ..:.:*:.: :.* . .*::*: :.
MMAR_2354|M.marinum_M VSERTND-DTRANAVTAITFDLDPDRARTDLQFIRNETKQSLSALKDTPN
MUL_1530|M.ulcerans_Agy99 VSERTND-DTRANAVTAITFDLDPDRARTDLQFIRNETKQSLSALKDTPN
MAV_4881|M.avium_104 VNERVAG-DTRANAVTNVDVLLGPAAIGTDLRAIRAAVKHALVRHHDLPD
MSMEG_4730|M.smegmatis_MC2_155 LNERTSLDDSRALAMTFASASIDPRQCTSDLTGVRATIREAIETARTTTD
MAB_4701|M.abscessus_ATCC_1997 ISDRTEN-DTRAMALSYARVSIDPVSLTTDLREVRAAIKQTLKTLKESPD
:.:*. *:** *:: :.* :** :* :.:: : .:
MMAR_2354|M.marinum_M ELLKPLPLVPYTPKWLARKMEV-LAMGTSDLPVGCSNLGELSQALTRVDG
MUL_1530|M.ulcerans_Agy99 ELLKPLPLVPYTPKWLARKMEV-LAMGTSDLPVGCSNLGELSQALTRVDG
MAV_4881|M.avium_104 ERWALLPLVPLIPKRVFRRLLG-VVTG-GATTVVSSNLGALDPAANRPDG
MSMEG_4730|M.smegmatis_MC2_155 ETLELLPLIPLVPKRALQGLGGLFFGSPDDLPVYCSNLGDVDSAVGRADG
MAB_4701|M.abscessus_ATCC_1997 ESKLLLWLPSFAPERTLKRMVARMPADPD-QSVFCSYLGDLHSMIGWPAG
* * * . *: : : . . . .* .* ** : *
MMAR_2354|M.marinum_M TGARYSSARMFNRGATRQRFERESGELYLFSGRLENSVFISVSAYQPGAE
MUL_1530|M.ulcerans_Agy99 TGARYSSARMFNRGATRQRFERESGELYLFSGRLENSVFISVSAYQPGAE
MAV_4881|M.avium_104 TDADYFAMRSVYPGVTDATMQRTNGALALLSGRVHDRVFVSVLGYELGRQ
MSMEG_4730|M.smegmatis_MC2_155 TDAEYVVMRGVDQNVTRRDIEAMGGQLVLVSVRVNGKISIGIVAYQLDAQ
MAB_4701|M.abscessus_ATCC_1997 TLADFQNARATGQRERRQLLERTGGRMFILSGRLNGKIFISVGAYQPGAT
* * : * :: .* : :.* *:.. : :.: .*: .
MMAR_2354|M.marinum_M NSNQYLRGLVEQTLADYSLKAELLG---------
MUL_1530|M.ulcerans_Agy99 NSNQYLRGLVEQTLADYSLKAELLG---------
MAV_4881|M.avium_104 NSNEALRQALSGALNDFSLTGATGWNPVRAVATR
MSMEG_4730|M.smegmatis_MC2_155 NSKTRLRELAALTLKDFGLAGVIE----------
MAB_4701|M.abscessus_ATCC_1997 NTKEALRELAELTLADFSLPGEIH----------
*:: ** :* *:.* .