For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TPEQMLDVAVEEARKGLSEGGIPIGAALFSADGELLGSGHNRRVQDGDPSIHAETDAFRAAGRQRGYRKT IMVTTLSPCWYCSGLVRQFNIGALVIGEARTFSGGHEWLADNGVSVTLLDDERCVTMMQDFITANPDLWA EDIGE*
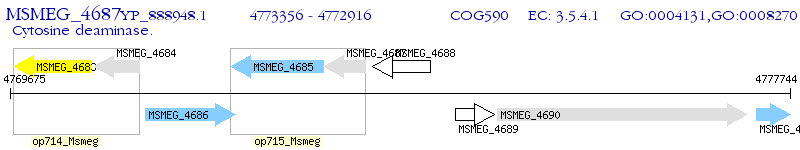
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4687 | - | - | 100% (146) | cytosine deaminase |
| M. smegmatis MC2 155 | MSMEG_6327 | - | 7e-11 | 34.78% (115) | cytidine and deoxycytidylate deaminase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3778c | - | 5e-08 | 34.41% (93) | cytidine/deoxycytidylate deaminase |
| M. gilvum PYR-GCK | Mflv_2579 | - | 1e-69 | 81.51% (146) | CMP/dCMP deaminase, zinc-binding |
| M. tuberculosis H37Rv | Rv3752c | - | 5e-08 | 34.41% (93) | cytidine/deoxycytidylate deaminase |
| M. leprae Br4923 | MLBr_02474 | - | 2e-09 | 34.21% (114) | putative cytidine/deoxycytidylate deaminase |
| M. abscessus ATCC 19977 | MAB_0268 | - | 4e-11 | 33.93% (112) | cytidine/deoxycytidylate deaminase family protein |
| M. marinum M | MMAR_5295 | - | 1e-09 | 37.63% (93) | cytidine/deoxycytidylate deaminase |
| M. avium 104 | MAV_0324 | - | 2e-08 | 36.56% (93) | cytidine and deoxycytidylate deaminase family protein |
| M. thermoresistible (build 8) | TH_1653 | - | 9e-12 | 35.65% (115) | POSSIBLE CYTIDINE/DEOXYCYTIDYLATE DEAMINASE |
| M. ulcerans Agy99 | MUL_4370 | - | 5e-10 | 37.63% (93) | cytidine/deoxycytidylate deaminase |
| M. vanbaalenii PYR-1 | Mvan_4055 | - | 5e-74 | 86.90% (145) | CMP/dCMP deaminase, zinc-binding |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3778c|M.bovis_AF2122/97 -----------------MTTDEDLIRAALAVAATAGPRD-VPVGAVVVGA
Rv3752c|M.tuberculosis_H37Rv -----------------MTTDEDLIRAALAVAATAGPRD-VPVGAVVVGA
MLBr_02474|M.leprae_Br4923 ----MPSAAGRRHQTRSVTTDEDLIRAALTVATTAGSRD-VPIGAVVLGA
MAV_0324|M.avium_104 -----------------MISDEDLIRSALAVAATAGPRD-VPIGAVVVGA
MMAR_5295|M.marinum_M -----------------MIADEDLVRAALAAAATAGPRD-VPIGAVVISA
MUL_4370|M.ulcerans_Agy99 -----------------MIADEDLVRAALAAAATAGPRD-VPIGAVMISA
TH_1653|M.thermoresistible__bu --------------MIGSVTDEDMIRAALTAAREAGPRD-VPIGAVVFGP
MAB_0268|M.abscessus_ATCC_1997 ---------------MTFPDDEALIRAALVAAREAGSQD-VPIGAVIYAA
Mflv_2579|M.gilvum_PYR-GCK ------------------MTASQMLDVAVEEARKGLAEGGIPIGAALFAA
Mvan_4055|M.vanbaalenii_PYR-1 MIRILGGVSLQDHRREGVATPQEMLEVAVEEARKGLAEGGIPIGAALFST
MSMEG_4687|M.smegmatis_MC2_155 ------------------MTPEQMLDVAVEEARKGLSEGGIPIGAALFSA
. :: *: * . ... :*:**.: ..
Mb3778c|M.bovis_AF2122/97 DGTELARAVNAREALGDPTAHAEILAMRLAAGVLGDGWRLEGTTLAVTVE
Rv3752c|M.tuberculosis_H37Rv DGTELARAVNAREALGDPTAHAEILAMRLAAGVLGDGWRLEGTTLAVTVE
MLBr_02474|M.leprae_Br4923 DGNELARAVNAREAIGDPTAHAEILAMRAAAGTLGNGWRLEGTTLAVTVE
MAV_0324|M.avium_104 DGTELARAVNAREALGDPTAHAEILALRAAAAALGDGWRLEGATLAVTVE
MMAR_5295|M.marinum_M DGTELARAVNAREELGDPTAHAEILALRAAARVLGDGWRLEGATLAVTVE
MUL_4370|M.ulcerans_Agy99 DGTELARAVNAREELGDPTAHAEILALRAAARVLGDGWRLEGATLAVTVE
TH_1653|M.thermoresistible__bu DGGELARAANAREQLGDPTAHAEVLALRAAARRLGDGWRLEGATLAVTVE
MAB_0268|M.abscessus_ATCC_1997 DGTELARAANARERLGDPTAHAEVLALRAAAAKHGDGWRLEGATLAVTVE
Mflv_2579|M.gilvum_PYR-GCK DGTLLGSGHNRRVQLDDPSLHAETDAFRNAGRQRG----YRSTTMVTTLS
Mvan_4055|M.vanbaalenii_PYR-1 DGALLGSGHNRRVQLGDPSVHAETDAFRNAGRQRG----YRSTIMVTTLS
MSMEG_4687|M.smegmatis_MC2_155 DGELLGSGHNRRVQDGDPSIHAETDAFRAAGRQRG----YRKTIMVTTLS
** *. . * * .**: *** *:* *. * . : :..*:.
Mb3778c|M.bovis_AF2122/97 PCTMCAGALVLARVARLVFGAWEPKTGAVGSLWDVVRDRRLNHRPEVRGG
Rv3752c|M.tuberculosis_H37Rv PCTMCAGALVLARVARLVFGAWEPKTGAVGSLWDVVRDRRLNHRPEVRGG
MLBr_02474|M.leprae_Br4923 PCTMCAGALVLARIERLVFGAWQPKTGAVGSLWDVVRDHRLNHRPAVRGG
MAV_0324|M.avium_104 PCTMCAGALVLARIARLVFGAWEPKTGAVGSLWDVVRDRRLNHRPEVRGG
MMAR_5295|M.marinum_M PCTMCAGALVLARVARLVFGAWEPKTGAVGSLWDVVRDRRLNHRPQVRGG
MUL_4370|M.ulcerans_Agy99 PCTMCAGALVLARVARLVFGAWEPKTGAVGSLWDVVRDRRLNHRPQVRGG
TH_1653|M.thermoresistible__bu PCTMCAGALVLARVARLVFGAWEPKTGAVGSLWDVVRDRRLNHRPEVRGG
MAB_0268|M.abscessus_ATCC_1997 PCTMCAGALVLARVGRVVFGAWEPKTGAVGSLWDVVRDRRQAHRPEVRGG
Mflv_2579|M.gilvum_PYR-GCK PCWYCSGLVRQFNIGTVVIGESRTFTG--GHDWLAEHGVKVTVVDDERCV
Mvan_4055|M.vanbaalenii_PYR-1 PCWYCSGLVRQFNIGAVVIGESRTFTG--GHDWLADNGVDVTIVDDERCV
MSMEG_4687|M.smegmatis_MC2_155 PCWYCSGLVRQFNIGALVIGEARTFSG--GHEWLADNGVSVTLLDDERCV
** *:* : .: :*:* .. :* * * . .. *
Mb3778c|M.bovis_AF2122/97 VLARECAAPLEAFFARQRLG-------
Rv3752c|M.tuberculosis_H37Rv VLARECAAPLEAFFARQRLG-------
MLBr_02474|M.leprae_Br4923 VLAQECTAPLEAFFAHQRLSKGPPVR-
MAV_0324|M.avium_104 VLERECAALLEGFFARQRLG-------
MMAR_5295|M.marinum_M VLAQECAAPLEEFFGRQRLG-------
MUL_4370|M.ulcerans_Agy99 VLAQECAAPLEEFFGRQRLG-------
TH_1653|M.thermoresistible__bu VLAAECAAPLEEFFARQRDPDSPRGLG
MAB_0268|M.abscessus_ATCC_1997 VLEHECAALLERFFAAKR---------
Mflv_2579|M.gilvum_PYR-GCK TLMRDFIAGNPELWAEDIGE-------
Mvan_4055|M.vanbaalenii_PYR-1 TMMRDFIAANPDLWAEDIGE-------
MSMEG_4687|M.smegmatis_MC2_155 TMMQDFITANPDLWAEDIGE-------
.: : : ::. .