For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PEGHTLHRLARLHQRRFGRTAVVVSSPQGRFADGAAAVSGQIFKRATAWGKHLFHHYDGGRVVHIHLGLY GAFTEWPVPAELALPLPVGQVRMRIIGAQYGTDLRGPTVCELITEPEIVDVIAKLGPDPLRPDADASLAW KRITKSRRPIGALLMDQTVMAGVGNVYRSELLFRHGIDPYLPGTQLDAAEFDAMWTDLVALMKVGVRRGK IVVVRPEHDHGAPSYRTGRPRTYVYRRAGEPCRICGTPVRTAELEGRNLFWCPTCQS*
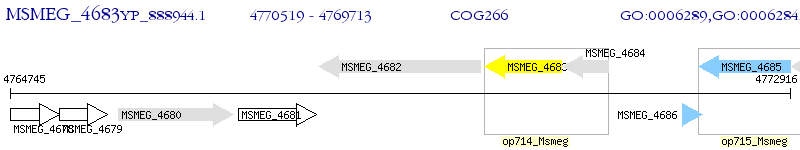
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4683 | - | - | 100% (268) | putative formamidopyrimidine-DNA glycosylase |
| M. smegmatis MC2 155 | MSMEG_1756 | - | 2e-23 | 32.84% (271) | endonuclease VIII and DNA n-glycosylase with an AP lyase |
| M. smegmatis MC2 155 | MSMEG_5545 | - | 2e-17 | 28.98% (283) | formamidopyrimidine-DNA glycosylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2491c | - | 1e-120 | 73.51% (268) | DNA glycosylase |
| M. gilvum PYR-GCK | Mflv_2582 | - | 1e-131 | 78.73% (268) | formamidopyrimidine-DNA glycolase |
| M. tuberculosis H37Rv | Rv2464c | - | 1e-120 | 73.51% (268) | DNA glycosylase |
| M. leprae Br4923 | MLBr_01658 | fpg | 6e-10 | 27.88% (226) | formamidopyrimidine-DNA glycosylase |
| M. abscessus ATCC 19977 | MAB_1575 | - | 1e-106 | 67.79% (267) | formamidopyrimidine-DNA glycolase |
| M. marinum M | MMAR_3812 | - | 1e-122 | 75.75% (268) | formamidopyrimidine-DNA glycosylase |
| M. avium 104 | MAV_1708 | - | 1e-124 | 75.66% (267) | endonuclease VIII and DNA n-glycosylase with an AP lyase |
| M. thermoresistible (build 8) | TH_0348 | - | 1e-120 | 73.41% (267) | POSSIBLE DNA GLYCOSYLASE |
| M. ulcerans Agy99 | MUL_3737 | - | 1e-120 | 75.37% (268) | formamidopyrimidine-DNA glycosylase |
| M. vanbaalenii PYR-1 | Mvan_4051 | - | 1e-133 | 78.73% (268) | formamidopyrimidine-DNA glycolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2582|M.gilvum_PYR-GCK MPEGHTLHRLAR-----QHQRRFGRAPVIVSSPQGRFVDGATAVNGRVLK
Mvan_4051|M.vanbaalenii_PYR-1 MPEGHTLHRLAR-----QHQRRFGRAPVIVSSPQGRFVDGAAAVNGHVFK
MSMEG_4683|M.smegmatis_MC2_155 MPEGHTLHRLAR-----LHQRRFGRTAVVVSSPQGRFADGAAAVSGQIFK
MMAR_3812|M.marinum_M MPEGHTLHRLAR-----LHQRRFAGAPVVVSSPQGKFADSAGAVDGRVLR
MUL_3737|M.ulcerans_Agy99 MPEGHTLHRLAR-----LHQRRFAGAPVVVSSPQGKFADSAGAVDGRVLR
MAV_1708|M.avium_104 MPEGHTLHRLAR-----LHQRRYAGAPVAVTSPQGRFAEAAAVVDGRVLR
Mb2491c|M.bovis_AF2122/97 MPEGHTLHRLAR-----LHQRRFAGAPVSVSSPQGRFADSASALNGRVLR
Rv2464c|M.tuberculosis_H37Rv MPEGHTLHRLAR-----LHQRRFAGAPVSVSSPQGRFADSASALNGRVLR
TH_0348|M.thermoresistible__bu MPEGHTLHRLAR-----LHERRFAGVRVRVTSPQGRFAEGAAAVSGRVFQ
MAB_1575|M.abscessus_ATCC_1997 MPEGHTLHRLAR-----LHQRRFAGAPVSVSSPQGRFTEGAAAVNGRTFV
MLBr_01658|M.leprae_Br4923 MPELPEVEVVRRGLQDYIVGKTITAVRVHHPRAVRRHVAGPTDLTNRLLG
*** :. : * : . * . . :.. .. : .: :
Mflv_2582|M.gilvum_PYR-GCK K----ATAWGKHLFHHYDGGRVVHVHLGLYGSFTEWSLP----PLLPVGQ
Mvan_4051|M.vanbaalenii_PYR-1 K----ASAWGKHLFHHYQGGRVVHVHLGLYGTFTEWPLPDDRTQPIPVGQ
MSMEG_4683|M.smegmatis_MC2_155 R----ATAWGKHLFHHYDGGRVVHIHLGLYGAFTEWPVPAELALPLPVGQ
MMAR_3812|M.marinum_M A----ASAWGKHLFHHYAGGPVVHVHLGLYGAFTEWARSAGELLPDPVGQ
MUL_3737|M.ulcerans_Agy99 A----ASAWGKHLFHHYAGGPVVHVHLGLYGAFTEWVRSAGELLPDPVGQ
MAV_1708|M.avium_104 R----TSAWGKHLFHHYAGGPIVHVHLGLYGSFSEWERPGDGPLPDPVGQ
Mb2491c|M.bovis_AF2122/97 R----ASAWGKHLFHHYVGGPVVHVHLGLYGTFTEWARPTDGWLPEPAGQ
Rv2464c|M.tuberculosis_H37Rv R----ASAWGKHLFHHYVGGPVVHVHLGLYGTFTEWARPTDGWLPEPAGQ
TH_0348|M.thermoresistible__bu R----SDAWGKHLFHHYEGGRVVHVHLGLYGTFTEFEIPDGAEPPPPVGQ
MAB_1575|M.abscessus_ATCC_1997 Q----AHAWGKHLFHDYGPVGVVHVHLGLYGAFTELPVPMG----LPVGQ
MLBr_01658|M.leprae_Br4923 TRINGIDRRGKYLWFLLDTDIALVVHLGMSGQMLLGTVPRVDHVRISALF
**:*:. : :***: * : . ..
Mflv_2582|M.gilvum_PYR-GCK VRMRMVGTEYGADLRGPTVCEVIDEPE--IADIVARLGPDPLRPDADPSL
Mvan_4051|M.vanbaalenii_PYR-1 VRMRMLGAEYGTDLRGPTVCEVIEEPD--VADVVARLGPDPLRSDADPEL
MSMEG_4683|M.smegmatis_MC2_155 VRMRIIGAQYGTDLRGPTVCELITEPE--IVDVIAKLGPDPLRPDADASL
MMAR_3812|M.marinum_M VRMRMVGAEYGTDLRGPTVCEVIDDAQ--VADVLARLGPDPLRKDADPSW
MUL_3737|M.ulcerans_Agy99 VRMRMVGAEYGTDLRGPTVCEVIDDAQ--VADVLARLGPDPLRKDADPSW
MAV_1708|M.avium_104 VRMRMVGASHGTDLRGPTVCEVIDEGQ--VSDVLARLGPDPLRDDADPSW
Mb2491c|M.bovis_AF2122/97 VRMRMVGAEFGTDLRGPTVCESIDDGE--VADVVARLGPDPLRSDANPSS
Rv2464c|M.tuberculosis_H37Rv VRMRMVGAEFGTDLRGPTVCESIDDGE--VADVVARLGPDPLRSDANPSS
TH_0348|M.thermoresistible__bu VRMRIIGGRYGTDLRGPTVCEVIGEPG--VDEIIARLGPDPLRPDADPDR
MAB_1575|M.abscessus_ATCC_1997 VRMRIEGAEFGTDLRGATACELIDAPQ--VDAILARLGPDPLRPRSDPAS
MLBr_01658|M.leprae_Br4923 DDGTVLNFTDQRTLGGWLLADLMTVDGSVLPVPVAHLARDPFDPRFDVEA
: . * * .: : : :*:*. **: :
Mflv_2582|M.gilvum_PYR-GCK AWRRISKSRRPIGALLMDQSVIAGIGNVYRSELLFRHRTDPFRPGTSVTA
Mvan_4051|M.vanbaalenii_PYR-1 AWRRISKSRRPIGALLMDQTVIAGVGNVYRSELLFRHRTDPFRPGTTVTS
MSMEG_4683|M.smegmatis_MC2_155 AWKRITKSRRPIGALLMDQTVMAGVGNVYRSELLFRHGIDPYLPGTQLDA
MMAR_3812|M.marinum_M AWARIAKSRRPIGALLMDQKVMAGVGNVYRSELLFRHRIDPYRSGQRITE
MUL_3737|M.ulcerans_Agy99 AWARIAKSRRPIGALLMDQKVMAGVGNVYRSELLFRHRIDPYRSGQRITE
MAV_1708|M.avium_104 AWQRIAKSRRPIGALLMDQTVMAGVGNVYRSELLFRHGIDPYRAGRDVGE
Mb2491c|M.bovis_AF2122/97 AWSRITKSRRPIGALLMDQTVIAGVGNVYRNELLFRHRIDPQRPGRGIGE
Rv2464c|M.tuberculosis_H37Rv AWSRITKSRRPIGALLMDQTVIAGVGNVYRNELLFRHRIDPQRPGRGIGE
TH_0348|M.thermoresistible__bu AWTRISNSRRAIGALLMDQKVIAGIGNVYRSELLFRHGIDPFRPGKQLEE
MAB_1575|M.abscessus_ATCC_1997 AFERIAKSHRPIGALLMDQKIIAGVGNVYRSEVLFRRRIDPYREGSRLDP
MLBr_01658|M.leprae_Br4923 VVKVLRCKHSELKRQLLDQQTVSGIGNIYADEALWRAEVHGARIAATLTR
. : .: : *:** ::*:**:* .* *:* . . :
Mflv_2582|M.gilvum_PYR-GCK DEFDALWTDLVALMKVGVRRGKIVVVLPEHDHGAPSYREGRPRTYVYRRA
Mvan_4051|M.vanbaalenii_PYR-1 DEFAEMWTDLVALMKVGVRRGKIVVVAPEHDHGAPSYREGRPRTYVYRRA
MSMEG_4683|M.smegmatis_MC2_155 AEFDAMWTDLVALMKVGVRRGKIVVVRPEHDHGAPSYRTGRPRTYVYRRA
MMAR_3812|M.marinum_M AEFSAAWTDLVALMKVGLRGGKIVVVRPEHDHGAPSYAAGRPRTYVYRRA
MUL_3737|M.ulcerans_Agy99 AEFSAAWTDLVALMKVGSRGGKIVVVRPEHDHGAPSYAAGRPRTYVYRRA
MAV_1708|M.avium_104 AEFDAAWTDLVALMKVGLRRGKIIVVRPEHDRGAPSYRPDRPRTYVYRRA
Mb2491c|M.bovis_AF2122/97 PEFDAAWNDLVSLMKVGLRRGKIIVVRPEHDHGLPSYLPDRPRTYVYRRA
Rv2464c|M.tuberculosis_H37Rv PEFDAAWNDLVSLMKVGLRRGKIIVVRPEHDHGLPSYLPDRPRTYVYRRA
TH_0348|M.thermoresistible__bu AEFCAMWRDLVELMKVGLRTGRIHVVRPEHDHGPPAYAPGRPRTYVYRRT
MAB_1575|M.abscessus_ATCC_1997 EQLTALWSDLVDRMRVGLRVGKIVTVDPEHDCGDPSYAPDRPRTYVYRRA
MLBr_01658|M.leprae_Br4923 RQLAAVLDAAADVMRDSLAKGGTSFDSLYVNVNGESGYFDRS-LDAYGRE
:: . *: . * : . : .*. .* *
Mflv_2582|M.gilvum_PYR-GCK GDPCRVCSTPVRTVEMEARNLFWCPNCQS----
Mvan_4051|M.vanbaalenii_PYR-1 GDPCRVCATPIRTVELEGRNLFWCPACQT----
MSMEG_4683|M.smegmatis_MC2_155 GEPCRICGTPVRTAELEGRNLFWCPTCQS----
MMAR_3812|M.marinum_M GDPCRVCGATVGTAVLEGRNVFWCPSCQA----
MUL_3737|M.ulcerans_Agy99 GDPCRVCGATVGTAVLEGRNVFWRPSCQA----
MAV_1708|M.avium_104 GEACRVCGEPVRTAVLEGRNVFWCPTCQK----
Mb2491c|M.bovis_AF2122/97 GEPCRVCGGVIRTALLEGRNVFWCPVCQT----
Rv2464c|M.tuberculosis_H37Rv GEPCRVCGGVIRTALLEGRNVFWCPVCQT----
TH_0348|M.thermoresistible__bu GEPCRVCGTAVSTAQLEARNVYWCPSCQR----
MAB_1575|M.abscessus_ATCC_1997 GAPCRVCGTPILTAEMDARNLFWCPSCQIG---
MLBr_01658|M.leprae_Br4923 GEGCRRCGAVMHREKFMNRSSFYCPRCQPRPRR
* ** *. : : *. :: * **