For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RLSRLVAAAGVGVLMLGASACSSTGGKPDSSGGGDMGAGTADTPRFTVAMITHEVPGDSFWDLVRKGAEA AAKKDNIELRYSSDPEAPNQANHVQSAVDSGVDGIAVTLAKPDAMKPAVQNATDKGIPVVAFNAGLDAWQ GMGVKEYFGQDGYIAGQEAGKRLAAEGAKKVICVIQEQGHVDLEARCAGVKNTFPASEVLNVNGKDMPSV ESTITAKLQQDPAIDTILTLGAPFALTAVQSAKNAGSQAKIATFDTNAALVDAIQNGDVQWAVDQQPFLQ GYLAVDSLWLYLNNGNVIGGNQPTLTGPSFIDKSNIDAVAEYAKNGTR*
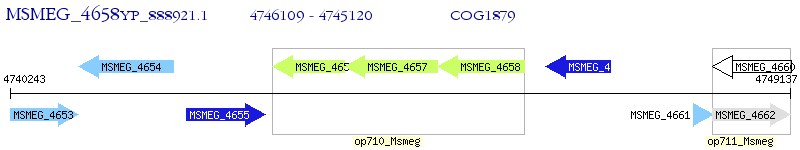
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4658 | - | - | 100% (329) | sugar ABC transporter substrate-binding protein |
| M. smegmatis MC2 155 | MSMEG_3599 | - | 3e-14 | 29.04% (272) | sugar-binding transcriptional regulator, LacI family protein |
| M. smegmatis MC2 155 | MSMEG_3598 | - | 5e-14 | 28.26% (276) | periplasmic sugar-binding proteins |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2605 | - | 1e-151 | 79.94% (329) | periplasmic binding protein/LacI transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4011 | - | 1e-156 | 82.67% (329) | periplasmic binding protein/LacI transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2605|M.gilvum_PYR-GCK MTFKRLAAIAGAGVLAFGVVSCSSTGGAPRESGEG-GGGGTVDTPRMTIA
Mvan_4011|M.vanbaalenii_PYR-1 MTFKRLAAFAGAGVLAMGIVSCSSTGGAPEQSGDG-GGGGTVDTPRMTIA
MSMEG_4658|M.smegmatis_MC2_155 MRLSRLVAAAGVGVLMLGASACSSTGGKPDSSGGGDMGAGTADTPRFTVA
* :.**.* **.*** :* :****** * .** * *.**.****:*:*
Mflv_2605|M.gilvum_PYR-GCK MITHEVPGDSFWDLVRKGAEAAAQKDNVELRYSNDPEAPNQANLVQAAID
Mvan_4011|M.vanbaalenii_PYR-1 MITHEVPGDSFWDLVRKGAEAAAKKDNIELRYSNDPEAPNQANLVQSAID
MSMEG_4658|M.smegmatis_MC2_155 MITHEVPGDSFWDLVRKGAEAAAKKDNIELRYSSDPEAPNQANHVQSAVD
***********************:***:*****.********* **:*:*
Mflv_2605|M.gilvum_PYR-GCK SNVDGIAVTLAKPDAMQAAVKAAEAKGIPVVAFNAGMDSWKAMGVKEYFG
Mvan_4011|M.vanbaalenii_PYR-1 SNVDGIAVTLAKPDAMQAAVKSAESKGIPVVAFNAGMDSWKAMGVKEYFG
MSMEG_4658|M.smegmatis_MC2_155 SGVDGIAVTLAKPDAMKPAVQNATDKGIPVVAFNAGLDAWQGMGVKEYFG
*.**************:.**: * ***********:*:*:.********
Mflv_2605|M.gilvum_PYR-GCK QDGRIAGQGVGDRLRAEGATKAVCIVQEQGHVDLEARCAGVRETFPATEV
Mvan_4011|M.vanbaalenii_PYR-1 QDGRIAGQGVGDRLRAEGATKSICVIQEQGHVDLEARCAGVKETFPATEI
MSMEG_4658|M.smegmatis_MC2_155 QDGYIAGQEAGKRLAAEGAKKVICVIQEQGHVDLEARCAGVKNTFPASEV
*** **** .*.** ****.* :*::***************::****:*:
Mflv_2605|M.gilvum_PYR-GCK LNVNGKDMPSVESTITAKLQQDPSIDYLVALGAPFALTAVQSAKNAGSTA
Mvan_4011|M.vanbaalenii_PYR-1 LNVNGKDMPSVESTITAKLQQDPSIDYVVTLGAPFALTAVQSAKNAGSSA
MSMEG_4658|M.smegmatis_MC2_155 LNVNGKDMPSVESTITAKLQQDPAIDTILTLGAPFALTAVQSAKNAGSQA
***********************:** :::****************** *
Mflv_2605|M.gilvum_PYR-GCK KIGTFDTNAALVSAIQAGDVQWAVDQQPYLQGYLAVDSLWLYLSNGNVIG
Mvan_4011|M.vanbaalenii_PYR-1 KIGTFDTNAALVEAIQNGDVQWAVDQQPYLQGYLAVDSLWLYLSNGNVIG
MSMEG_4658|M.smegmatis_MC2_155 KIATFDTNAALVDAIQNGDVQWAVDQQPFLQGYLAVDSLWLYLNNGNVIG
**.*********.*** ***********:**************.******
Mflv_2605|M.gilvum_PYR-GCK GGQPTLTGPAFIDQSNIDAVAEYAKAGTR
Mvan_4011|M.vanbaalenii_PYR-1 GGQPTLTGPAFIDKSNIDAVAEYAKGGTR
MSMEG_4658|M.smegmatis_MC2_155 GNQPTLTGPSFIDKSNIDAVAEYAKNGTR
*.*******:***:*********** ***