For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RLGTTAFAIASATALGLGLTACGAGDPAANSDTTRIGVTVYDMSSFITAGKEGMDAYAKDNNIELIWNSA NLDVSTQASQVDSMINQGVDAIIVVPVQADSLAPQVASAKAKGIPLVPVNAALDSKDIAGNVQPDDVAAG AQEMQMMADRLGGKGNIVILQGPLGQSGELDRSKGIEQVLAKYPDIKVLAKDTANWKRDEAVNKMKNWIS GFGPQIDGVVAQNDDMGLGALQALKESGRTGVPIVGIDGIEDGLNAVKSGDFIGTSLQNGTVELAAGLAV ANRLAKGEPVNKEPVYIMPAITKDNVDVAIEHVVTERQQFLDGLTELINKNLETGDIAYEGIPGQKQP*
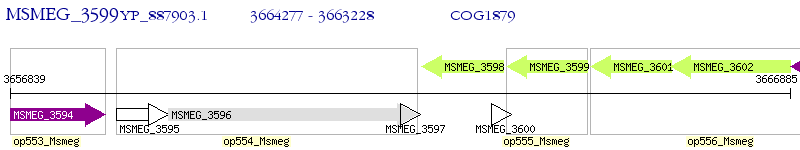
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3599 | - | - | 100% (349) | sugar-binding transcriptional regulator, LacI family protein |
| M. smegmatis MC2 155 | MSMEG_3598 | - | e-170 | 85.96% (349) | periplasmic sugar-binding proteins |
| M. smegmatis MC2 155 | MSMEG_3095 | - | 1e-31 | 30.88% (285) | D-ribose-binding periplasmic protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2605 | - | 2e-14 | 26.23% (324) | periplasmic binding protein/LacI transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00398 | - | 2e-08 | 24.69% (324) | putative D-ribose-binding protein |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5293 | - | 8e-22 | 25.15% (330) | solute-binding protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3599|M.smegmatis_MC2_155 ---------MRLGTTAFAIASATALGLGLTACGAGDPAANSDTT--RIGV
Mvan_5293|M.vanbaalenii_PYR-1 ---------MKR-TSSLLFTAVVGVGLTLTACGSNSDSGSTEARSGKVGV
Mflv_2605|M.gilvum_PYR-GCK MTFKRLAAIAGAGVLAFGVVSCSSTGGAPRESGEGGGGGTVDTPRMTIAM
MLBr_00398|M.leprae_Br4923 ---------MSTGTSVPLRVASIMLILGLLPAILPACGFSESHHKLTIGV
. .: . . . . :.:
MSMEG_3599|M.smegmatis_MC2_155 TVYDMSS---FITAGKEGMDAYAKDNNIELIWNSANLDVSTQASQVDSMI
Mvan_5293|M.vanbaalenii_PYR-1 ILPDTKSSVRWETKDRPALAAAFQNAGVDYTIQNAEGSADTMATIADGMI
Mflv_2605|M.gilvum_PYR-GCK ITHEVPGD-SFWDLVRKGAEAAAQKDNVELRYS-NDPEAPNQANLVQAAI
MLBr_00398|M.leprae_Br4923 SYPTANSP--FWNAYTRFIDEGSHQLGVYINAVSAEEDEQKQLSDVETLI
. : :. .: : . . . .: *
MSMEG_3599|M.smegmatis_MC2_155 NQGVDAIIVVPVQADSLAPQVASAKAKGIPLVPVN---AALDSKDIAGNV
Mvan_5293|M.vanbaalenii_PYR-1 ADGVTVLAIVNLDSDSGASIQQKAASQGVKTIDYD---RLTLGGSADVYV
Mflv_2605|M.gilvum_PYR-GCK DSNVDGIAVTLAKPDAMQAAVKAAEAKGIPVVAFN---AGMDSWKAMG-V
MLBr_00398|M.leprae_Br4923 NQGIDGLIVTPQSTSIAPTLLRIATQANIPVVVVDRYPGYAPGQNKNADY
..: : :. ... . * .: : : . .
MSMEG_3599|M.smegmatis_MC2_155 QPDDVAAGAQEMQMMADRLGGK-GNIVILQGPLGQSGELDRSKGIEQVLA
Mvan_5293|M.vanbaalenii_PYR-1 SFDNNVVGELQGQGLVDCLGGRPANVVFLNGSPTDNNATLFSSGAHSVVD
Mflv_2605|M.gilvum_PYR-GCK KEYFGQDGRIAGQGVGDRLRAEGATKAVCIVQ--EQGHVDLEARCAGVRE
MLBr_00398|M.leprae_Br4923 VAFLGPNNEKAGSGIAEALMAGGGSKFLALGGMPGNSVAQGRKAGLESAL
. . : : * . .. . ..
MSMEG_3599|M.smegmatis_MC2_155 KYPDIKVLAKDTA-NWKRDEAVNKMKNWISGFGPQIDGVVAQNDDMGLGA
Mvan_5293|M.vanbaalenii_PYR-1 ATPSITIVGEQAVPDWDNDKAVTIFEQLYTAADGRVDGVYAANDGLAGSV
Mflv_2605|M.gilvum_PYR-GCK TFPATEVLNVNGK----DMPSVESTITAKLQQDPSIDYLVALGAPFALTA
MLBr_00398|M.leprae_Br4923 TAVGHRLVQFQYAGDSEDKGLAAAENMLQAHPAGDANAIWCFNDKLCQGA
:: : . : : . . : .
MSMEG_3599|M.smegmatis_MC2_155 LQALKESGRTG-VPIVGIDGIEDGLNAVKSGDFIGTSLQNGTVELAAGLA
Mvan_5293|M.vanbaalenii_PYR-1 ISILEKNKRAGQVPVTGQDATVEGLQNILAGTQCMTVYKSATEEANALAE
Mflv_2605|M.gilvum_PYR-GCK VQSAKNAGSTA--KIGTFDTNAALVSAIQAGDVQWAVDQQPYLQGYLAVD
MLBr_00398|M.leprae_Br4923 IKAVYNANREKEFIFGGMDLTPQAIAAIENG-LYTVSLGAHWLEGGFGLA
:. : . * : : * . :
MSMEG_3599|M.smegmatis_MC2_155 VANRLAKGEPVNKEPVYIMPAITKDNVDVAIEHVVTERQQFLDGLTELIN
Mvan_5293|M.vanbaalenii_PYR-1 VAIALANGE----QPQTTSTSRDDTGGRDVPSVLLTPKSITKDNINVVFD
Mflv_2605|M.gilvum_PYR-GCK S------------LWLYLSNGNVIGGGQPTLTGPAFIDQSNIDAVAEYAK
MLBr_00398|M.leprae_Br4923 ILYRKIHGQ------DPAERMVKLDLLKVDKSNVAKFKARYIDNTPHYNW
*
MSMEG_3599|M.smegmatis_MC2_155 KNLETGDIAYEGIPGQKQP----
Mvan_5293|M.vanbaalenii_PYR-1 DGGQSKDEVCSGQFAEACSAAGV
Mflv_2605|M.gilvum_PYR-GCK AGTR-------------------
MLBr_00398|M.leprae_Br4923 KSTTPFDMTITLN----------
.