For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTDVTTSTENELRELAERGAAELADASAEELLRWTDEHFGGNYVVASNMQDAVLVEMAAKVRPGVDVLFL DTGYHFAETIGTRDAVEAVYDVHVVNVTPERTVAEQDELLGKNLFARDPGECCRLRKVVPLTNALKGYSA WVTGIRRVEAPTRANAPLISWDNAFGLVKINPIAAWTDEDMQNYIDANGILVNPLVYEGYPSIGCAPCTS KPIPGADPRSGRWAGLSKTECGLHVS
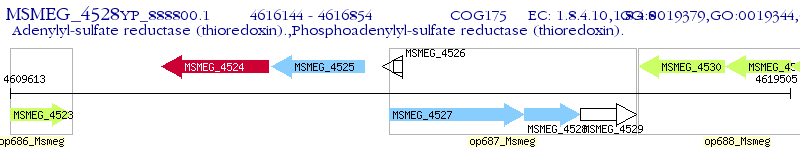
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4528 | - | - | 100% (236) | phosphoadenosine phosphosulfate reductase |
| M. smegmatis MC2 155 | MSMEG_4979 | cysD | 1e-05 | 23.30% (206) | sulfate adenylyltransferase subunit 2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2413 | cysH | 1e-102 | 70.92% (251) | phosphoadenosine phosphosulfate reductase |
| M. gilvum PYR-GCK | Mflv_2685 | - | 1e-109 | 79.32% (237) | phosphoadenosine phosphosulfate reductase |
| M. tuberculosis H37Rv | Rv2392 | cysH | 1e-102 | 70.92% (251) | phosphoadenosine phosphosulfate reductase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1661c | - | 1e-112 | 81.20% (234) | phosphoadenosine phosphosulfate reductase (CysH) |
| M. marinum M | MMAR_3417 | cysH_1 | 1e-104 | 73.53% (238) | 3'-phosphoadenosine 5'-phosphosulfate reductase CysH_1 |
| M. avium 104 | MAV_1786 | - | 1e-103 | 71.66% (247) | phosphoadenosine phosphosulfate reductase |
| M. thermoresistible (build 8) | TH_1444 | cysH | 1e-109 | 79.08% (239) | PROBABLE 3'-PHOSPHOADENOSINE 5'-PHOSPHOSULFATE REDUCTASE |
| M. ulcerans Agy99 | MUL_2696 | cysH_1 | 1e-105 | 73.95% (238) | phosphoadenosine phosphosulfate reductase |
| M. vanbaalenii PYR-1 | Mvan_3858 | - | 1e-114 | 84.78% (230) | phosphoadenosine phosphosulfate reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2685|M.gilvum_PYR-GCK MT-DTMS--ETDLQELAERGAAELGEHATAQDLLRWTDENFGP-------
Mvan_3858|M.vanbaalenii_PYR-1 ----MYT--EQDLRQLAERGAAELGEHATAEELLRWTDEHFG--------
TH_1444|M.thermoresistible__bu MT-DLMT--ETDLRALAERGAAEMAG-AGAEELLRWTDEHFGG-------
MSMEG_4528|M.smegmatis_MC2_155 MT-DVTTSTENELRELAERGAAELAD-ASAEELLRWTDEHFG--------
MAB_1661c|M.abscessus_ATCC_199 MSRETTTKDLDSLRALAERGAAELQD-ASAEELLRWTDENFG--------
Mb2413|M.bovis_AF2122/97 MSGETTRLTEPQLRELAARGAAELDG-ATATDMLRWTDETFGDIGGAGGG
Rv2392|M.tuberculosis_H37Rv MSGETTRLTEPQLRELAARGAAELDG-ATATDMLRWTDETFGDIGGAGGG
MAV_1786|M.avium_104 MTERTTKLPEAELRELAARGAAELEG-ASASDVLRWTDETFG-------G
MMAR_3417|M.marinum_M MSDQPN---ETQLRELAARGAAEMEG-ASATDLLRWADEHFG-------G
MUL_2696|M.ulcerans_Agy99 MSDQPN---ETQLRELAARGAAEMEG-ASATDLLRWADEHFG-------G
.*: ** *****: * * ::***:** **
Mflv_2685|M.gilvum_PYR-GCK ASG-----RGDYIVASNMQDAVLVDLAAKVRPGVDVLFLDTGYHFVETIG
Mvan_3858|M.vanbaalenii_PYR-1 ---------GDYIVASNMQDAVLVDLAAKVRPGVDVLFLDTGYHFVETIG
TH_1444|M.thermoresistible__bu APAGSAGARSGYVVASNMQDAVLIHLAAKVRPGVDVLFLDTGYHFAETIG
MSMEG_4528|M.smegmatis_MC2_155 ---------GNYVVASNMQDAVLVEMAAKVRPGVDVLFLDTGYHFAETIG
MAB_1661c|M.abscessus_ATCC_199 ---------GNYVVASNMQDAVLVDLAAQVRPGVDVLFLDTGYHFAETIG
Mb2413|M.bovis_AF2122/97 VSGHRGWTTCNYVVASNMADAVLVDLAAKVRPGVPVIFLDTGYHFVETIG
Rv2392|M.tuberculosis_H37Rv VSGHRGWTTCNYVVASNMADAVLVDLAAKVRPGVPVIFLDTGYHFVETIG
MAV_1786|M.avium_104 VNGPRGWATCNYVVASSMQEAVLIDLAAKVRPGVPVVFLDTGYHFAETIG
MMAR_3417|M.marinum_M INGPRGWATCNYVVASNMQDAVLIDLASKVRPGVPVIFLDTGYHFAETIG
MUL_2696|M.ulcerans_Agy99 INGPRGWATCNYVVASNMQDAVLIDLASKVRPGVPVIFLDTGYHFAETIG
.*:***.* :***:.:*::***** *:********.****
Mflv_2685|M.gilvum_PYR-GCK TRDAVEAVYDVNVVNVRPEHSVTEQDAMLGRNLFERNANECCRLRKVVPL
Mvan_3858|M.vanbaalenii_PYR-1 TRDAVEAVYDVNVVNVRPEHSVAEQDALLGKNLFERNANECCRLRKVEPL
TH_1444|M.thermoresistible__bu TRDAVQAVYDVNVVNVTAEHTVAEQDRLLGKDLFARDPNECCRLRKVEPL
MSMEG_4528|M.smegmatis_MC2_155 TRDAVEAVYDVHVVNVTPERTVAEQDELLGKNLFARDPGECCRLRKVVPL
MAB_1661c|M.abscessus_ATCC_199 TRDAVESVYDIHVVNVAPEQTVAQQDELVGKDLFASDPGECCRLRKVVPL
Mb2413|M.bovis_AF2122/97 TRDAIESVYDVRVLNVTPEHTVAEQDELLGKDLFARNPHECCRLRKVVPL
Rv2392|M.tuberculosis_H37Rv TRDAIESVYDVRVLNVTPEHTVAEQDELLGKDLFARNPHECCRLRKVVPL
MAV_1786|M.avium_104 TRDAIESVYDIRVLNVTPEHSVAEQDKLLGKDLFARDPGECCRLRKVAPL
MMAR_3417|M.marinum_M TRDAIEAVYDIRVLNISPEHTVAEQDKLLGKDLFARDPNECCRLRKVVPL
MUL_2696|M.ulcerans_Agy99 TRDAIEAVYDIRVLNISPEHTVAEQDKLLGKDLFARDPNECCRLRKVVPL
****:::***:.*:*: .*::*::** ::*::** :. ******** **
Mflv_2685|M.gilvum_PYR-GCK SKALSGYSAWVTGIRRVEAPTRANAPLISWDKAFGLVKINPLAAWSDDDM
Mvan_3858|M.vanbaalenii_PYR-1 SKALRGYSAWVTGIRRVEAPTRANAPLISWDSAFGLVKINPLAAWSDDDM
TH_1444|M.thermoresistible__bu SRALRGYSAWVTGIRRVEAPTRANAPFISFDESFGLVKINPLAAWTDQDM
MSMEG_4528|M.smegmatis_MC2_155 TNALKGYSAWVTGIRRVEAPTRANAPLISWDNAFGLVKINPIAAWTDEDM
MAB_1661c|M.abscessus_ATCC_199 RKSLAGYAAWVTGLRRVDAPTRANAPLISFDEAFGLVKINAIAAWTDEDM
Mb2413|M.bovis_AF2122/97 GKTLRGYSAWVTGLRRVDAPTRANAPLVSFDETFKLVKVNPLAAWTDQDV
Rv2392|M.tuberculosis_H37Rv GKTLRGYSAWVTGLRRVDAPTRANAPLVSFDETFKLVKVNPLAAWTDQDV
MAV_1786|M.avium_104 GKTLRGYSAWVTGLRRSEAATRANAPVIGFDEGFKLVKVNPMATWTDEDV
MMAR_3417|M.marinum_M TNALRGYAAWITGLRRVESPTRANAPVISFDEAFKLVKINPLVSWTDQDV
MUL_2696|M.ulcerans_Agy99 TNALRGYAAWITGLRRVESPTRANAPVISFDEAFKLVKINPLVAWTDQDV
.:* **:**:**:** ::.******.:.:*. * ***:*.:.:*:*:*:
Mflv_2685|M.gilvum_PYR-GCK QDYIDANGILVNPLVDEGYPSIGCAPCTHKPVEGADPRSGRWLGQSKTEC
Mvan_3858|M.vanbaalenii_PYR-1 QDYIDANGILVNPLVDEGYPSIGCAPCTNKPVEGADPRSGRWAGLSKTEC
TH_1444|M.thermoresistible__bu QDYIQTHGVLVNPLVDEGYPSIGCAPCTTKPAPGADPRSGRWQGTGKIEC
MSMEG_4528|M.smegmatis_MC2_155 QNYIDANGILVNPLVYEGYPSIGCAPCTSKPIPGADPRSGRWAGLSKTEC
MAB_1661c|M.abscessus_ATCC_199 QKYIDEHGTLVNPLVDEGYPSIGCAPCTAKPVLGGDARSGRWQGLAKTEC
Mb2413|M.bovis_AF2122/97 QEYIADNDVLVNPLVREGYPSIGCAPCTAKPAEGADPRSGRWQGLAKTEC
Rv2392|M.tuberculosis_H37Rv QEYIADNDVLVNPLVREGYPSIGCAPCTAKPAEGADPRSGRWQGLAKTEC
MAV_1786|M.avium_104 QNYIDEHNVLVNPLIYEGYSSIGCAPCTAKPLAGADPRSGRWQGLAKTEC
MMAR_3417|M.marinum_M QDYIQSNNVLVNPLVDEGYPSIGCAPCTAKPAVGEDPRSGRWRGRTKTEC
MUL_2696|M.ulcerans_Agy99 QDYIQSNNVLVNPLVDEGYPSIGCAPCTAKPAVGEDPRSGRWRGRTKTEC
*.** :. *****: ***.******** ** * *.***** * * **
Mflv_2685|M.gilvum_PYR-GCK GLHAS
Mvan_3858|M.vanbaalenii_PYR-1 GLHAS
TH_1444|M.thermoresistible__bu GLHSS
MSMEG_4528|M.smegmatis_MC2_155 GLHVS
MAB_1661c|M.abscessus_ATCC_199 GLHAS
Mb2413|M.bovis_AF2122/97 GLHAS
Rv2392|M.tuberculosis_H37Rv GLHAS
MAV_1786|M.avium_104 GLHAS
MMAR_3417|M.marinum_M GLHAS
MUL_2696|M.ulcerans_Agy99 GLHAS
*** *