For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSDHPVIDLSVDELLTTTRSVRRRLDLTRPVPLGLIRECLEIAVQAPTSGNLQNASFIVVTDGRQRRDIA DVYLSTWNQIAASPYAVADRFKDDPVRNRQQRRVSDSAAYYTGTTFHRAKRIPLDDVLHVDRW
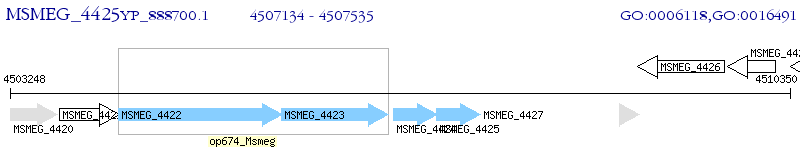
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4425 | - | - | 100% (133) | putative oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_1635 | - | 1e-19 | 44.34% (106) | nitroreductase family protein |
| M. smegmatis MC2 155 | MSMEG_1511 | - | 1e-14 | 50.75% (67) | putative oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3403c | - | 5e-21 | 45.13% (113) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_4892 | - | 4e-21 | 48.08% (104) | nitroreductase |
| M. tuberculosis H37Rv | Rv3368c | - | 5e-21 | 45.13% (113) | oxidoreductase |
| M. leprae Br4923 | MLBr_00418 | - | 2e-19 | 44.12% (102) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_3693c | - | 1e-19 | 42.31% (104) | oxidoreductase |
| M. marinum M | MMAR_1160 | - | 6e-18 | 41.35% (104) | nitroreductase |
| M. avium 104 | MAV_4334 | - | 1e-19 | 43.27% (104) | nitroreductase family protein |
| M. thermoresistible (build 8) | TH_2121 | - | 6e-19 | 44.23% (104) | POSSIBLE OXIDOREDUCTASE |
| M. ulcerans Agy99 | MUL_2854 | - | 1e-18 | 42.31% (104) | nitroreductase |
| M. vanbaalenii PYR-1 | Mvan_1532 | - | 8e-20 | 44.23% (104) | nitroreductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3403c|M.bovis_AF2122/97 ----MTLNLSVDEVLTTTRSVRKRLDFDKPVPRDVLMECLELALQAPTGS
Rv3368c|M.tuberculosis_H37Rv ----MTLNLSVDEVLTTTRSVRKRLDFDKPVPRDVLMECLELALQAPTGS
MMAR_1160|M.marinum_M ----MTLNLSVDEVLTTTRSVRKRLDFDKPVSREVLMECLDLALQAPTGS
MUL_2854|M.ulcerans_Agy99 ----MTLNLSVDEVLTTTRSVRKRLDFDKPVSREVLMECLDLALQAPTGS
MLBr_00418|M.leprae_Br4923 --------MSVDEVLTTTRSVRKRLDFDKPVPRDVLMECLQLALQAPTGS
MAV_4334|M.avium_104 ----MTLNLSVDEVLTTTRSVRKRLDFDKPVPREVLMECLELALQAPTGS
TH_2121|M.thermoresistible__bu ----MALNLSVDDVLTTTRSVRKRLDLEKPVPREVLMECLELALQAPTGS
Mflv_4892|M.gilvum_PYR-GCK ----MTLNLSADEVLTTTRSVRKRLDFDRPVPREVLNECLEIALQAPTGS
Mvan_1532|M.vanbaalenii_PYR-1 ----MTLNLSADEVLTTTRSVRKRLDFDKPVPRDVLMECLDIALQAPTGS
MAB_3693c|M.abscessus_ATCC_199 ---MTSLNLSADEVLTTTRSVRKRLDFDKPVERAVVEECLNIAMQAPTGS
MSMEG_4425|M.smegmatis_MC2_155 MSDHPVIDLSVDELLTTTRSVRRRLDLTRPVPLGLIRECLEIAVQAPTSG
:*.*::********:***: :** :: ***::*:****..
Mb3403c|M.bovis_AF2122/97 NSQGWQWVFVEDAAKKKAIADVYLANARGYLSGPAPEYPDGDTRGERMGR
Rv3368c|M.tuberculosis_H37Rv NSQGWQWVFVEDAAKKKAIADVYLANARGYLSGPAPEYPDGDTRGERMGR
MMAR_1160|M.marinum_M NSQGWQWVFVEDADKKKAIAEVYLARARSYLSMPAPEYAEGDTRGERMGK
MUL_2854|M.ulcerans_Agy99 NSQGWQWVFVEDADKKKAIADVYLARARSYLSMPAPEYAEGDTRGERMGK
MLBr_00418|M.leprae_Br4923 NSQGWHWVFVEDAEKRKAIGDIYLVNARCYLSQPAPEYPEGDTRGERMRL
MAV_4334|M.avium_104 NAQGWQWVFVEDADKKKAIADIYLANARAYLSLPAAEYPEGDTRGERMPK
TH_2121|M.thermoresistible__bu NAQGWQWVFVEDPDTKKALADIYRVNADAYLDRPRPSY--DDVRGERMDR
Mflv_4892|M.gilvum_PYR-GCK NAQGWQFVFVDDPEKKKALADIYRIAADPYLEAEKPTF--GDVRDERTPL
Mvan_1532|M.vanbaalenii_PYR-1 NNQGWQWVFVEDPEKKKAIADIYRTNATPYLAMEKPAT--GDMRDGQRPR
MAB_3693c|M.abscessus_ATCC_199 NHQGWHWVIVEDAEKKKAIADIYREGWNKYAKGAGATYAEGDTRAERKAK
MSMEG_4425|M.smegmatis_MC2_155 NLQNASFIVVTDGRQRRDIADVYLSTWN---------------------Q
* *. ::.* * :: :.::*
Mb3403c|M.bovis_AF2122/97 VRDSATYLAEHMHRAPVLLIPCLKGREDESAVG-GVSFWASLFPAVWSFC
Rv3368c|M.tuberculosis_H37Rv VRDSATYLAEHMHRAPVLLIPCLKGREDESAVG-GVSFWASLFPAVWSFC
MMAR_1160|M.marinum_M VRDSATYLAEHMHEAPVLMIPCIQGREDKSPLG-GVSFWASLFPAVWSFC
MUL_2854|M.ulcerans_Agy99 VRDSATYLAEHMHEAPVLMIPCIQGREDKSPLG-GVSLWASLFPAVWSFC
MLBr_00418|M.leprae_Br4923 VRDSATYLAEHMHEVPVLLIPCLLGRAEESPLG-AVSYWASLFPAVWSFC
MAV_4334|M.avium_104 VKDSAVYLAEHMHEAPVLLIPCLEGRVENAPLGLSASFWASLFPAAWSFC
TH_2121|M.thermoresistible__bu VKSSARYLSEHFHEVPVLLVPCLEGRPDNQPAGVTASFWGSLLPAVWSFM
Mflv_4892|M.gilvum_PYR-GCK VVDSAKYLAEHLHEAPVMLIPCLEGRPDGAPAGMSAGFWGSLLPAVWSFM
Mvan_1532|M.vanbaalenii_PYR-1 VEDSARYLNEHLEKVPVMLIPCLQGKPQDAVSGASAGYWGSLLPAVWSFM
MAB_3693c|M.abscessus_ATCC_199 VVDSAGYLAENFERSPLFLIPCIEGRLDNLPVVGAASSWGSLLPAVWSFM
MSMEG_4425|M.smegmatis_MC2_155 IAASPYAVADRFKDDPVRNR------------------------------
: *. : :.:. *:
Mb3403c|M.bovis_AF2122/97 LALRSRGLGSCWTTLHLLDNGEHKVADVLGIPYDEYSQGGLLPIAYTQGI
Rv3368c|M.tuberculosis_H37Rv LALRSRGLGSCWTTLHLLDNGEHKVADVLGIPYDEYSQGGLLPIAYTQGI
MMAR_1160|M.marinum_M LALRSRGLGSCWTTLHLLADGERQVADVLGIPYDDYSQGGLFPIAYTKGT
MUL_2854|M.ulcerans_Agy99 LALRSRGLGSCWTTLHLLADGERQVADVLGIPYDDYSQGGLFPIAYTKGT
MLBr_00418|M.leprae_Br4923 LALRSRGLGTCWTSLHLLGDGEQRAAEVLGIPSDKYSQGGLFPIAYTKGT
MAV_4334|M.avium_104 LALRSRGLGTCWTTLHLINDGEKQAAELLGIPHDRYSQGGLFPIAYTKGT
TH_2121|M.thermoresistible__bu LALRSRGLGSAWTTLHLVGDGEQRAAEVLGIPFDRYSQAGLFPIAYTKGT
Mflv_4892|M.gilvum_PYR-GCK LALRSRGLGSAWTTLHLIGDGEKQAAEVLGIPFDKYSQGGLFPIAYTKGT
Mvan_1532|M.vanbaalenii_PYR-1 LALRSRGLGSAWTTLHLLGDGERQAAEILGIPFDNYSQAGLFPIAYTKGT
MAB_3693c|M.abscessus_ATCC_199 LAARNRGLGSAWTTLHLMDDGEKRTADIVGIPFDKVTQGGLFPIAYTVGT
MSMEG_4425|M.smegmatis_MC2_155 --------------------QQRRVSDSA---------------AYYTGT
:::.:: ** *
Mb3403c|M.bovis_AF2122/97 DFRPAKRLPAESVTHWNGW
Rv3368c|M.tuberculosis_H37Rv DFRPAKRLPAESVTHWNGW
MMAR_1160|M.marinum_M DFRPAKRLPADRLTHWNSW
MUL_2854|M.ulcerans_Agy99 DFRPAKRLPADRLTHWNSW
MLBr_00418|M.leprae_Br4923 DFRPANRLPAENVTHWDIW
MAV_4334|M.avium_104 DFRPAKRLPAEQVTHWDSW
TH_2121|M.thermoresistible__bu DFRPAKRLPAEQVVHWNRW
Mflv_4892|M.gilvum_PYR-GCK DFKKAKRLPAEQLTHWNTW
Mvan_1532|M.vanbaalenii_PYR-1 DFRRAKRLPAEQLTHWDTW
MAB_3693c|M.abscessus_ATCC_199 DFRLAKRQPLADVLHWDTW
MSMEG_4425|M.smegmatis_MC2_155 TFHRAKRIPLDDVLHVDRW
*: *:* * : * : *