For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTHTAVLAGGCFWGLEDLIRALPGVISTRVGYTGGDNDHPTYRNHPGHAEAVEITYDPQQTDYRALLEFF FQIHDPTTKNRQGNDVGSSYRSAIFYLDDDQKRVAFDIVAEIDASGRWPGPVATEVTPAGVFWEAEPEHQ DYLQRNPGGYTCHWVRPEWILDPEADPETASDSGSDVSAASATASHHD*
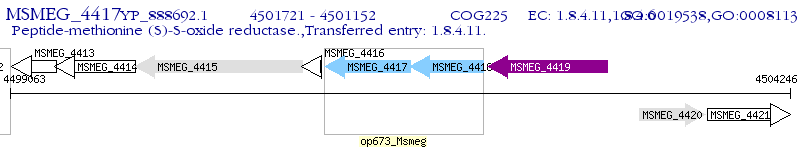
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4417 | msrA | - | 100% (189) | methionine sulfoxide reductase A |
| M. smegmatis MC2 155 | MSMEG_6477 | msrA | 1e-77 | 77.38% (168) | methionine sulfoxide reductase A |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0142c | msrA | 4e-70 | 71.69% (166) | methionine sulfoxide reductase A |
| M. gilvum PYR-GCK | Mflv_1105 | - | 2e-76 | 75.31% (162) | methionine sulfoxide reductase A |
| M. tuberculosis H37Rv | Rv0137c | msrA | 4e-70 | 71.69% (166) | methionine sulfoxide reductase A |
| M. leprae Br4923 | MLBr_02647 | - | 1e-69 | 68.60% (172) | methionine sulfoxide reductase A |
| M. abscessus ATCC 19977 | MAB_0120 | - | 2e-27 | 41.25% (160) | peptide methionine sulfoxide reductase |
| M. marinum M | MMAR_0347 | msrA | 2e-71 | 73.91% (161) | peptide methionine sulfoxide reductase MsrA |
| M. avium 104 | MAV_5159 | msrA | 2e-71 | 71.95% (164) | methionine sulfoxide reductase A |
| M. thermoresistible (build 8) | TH_0081 | - | 6e-79 | 79.62% (157) | PROBABLE PEPTIDE METHIONINE SULFOXIDE REDUCTASE MSRA |
| M. ulcerans Agy99 | MUL_4773 | msrA | 8e-71 | 73.29% (161) | methionine sulfoxide reductase A |
| M. vanbaalenii PYR-1 | Mvan_5709 | - | 2e-77 | 76.54% (162) | methionine sulfoxide reductase A |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0142c|M.bovis_AF2122/97 -------------------------MTSNQKAILAG--------------
Rv0137c|M.tuberculosis_H37Rv -------------------------MTSNQKAILAG--------------
MMAR_0347|M.marinum_M -------------------------MTSPQKAILAG--------------
MUL_4773|M.ulcerans_Agy99 -------------------------MTSPQKAILAG--------------
MAV_5159|M.avium_104 -------------------------MTNTQKAILAG--------------
MLBr_02647|M.leprae_Br4923 -----------------------MARTNTRKAILAG--------------
Mflv_1105|M.gilvum_PYR-GCK -------------------------MSDHKRAVLAG--------------
Mvan_5709|M.vanbaalenii_PYR-1 -------------------------MSDHKRAVLAG--------------
TH_0081|M.thermoresistible__bu -------------------------VSEYKKAILAG--------------
MSMEG_4417|M.smegmatis_MC2_155 --------------------------MTTHTAVLAG--------------
MAB_0120|M.abscessus_ATCC_1997 MASWVDNVRDQILGRIEDGLKPGKSAIVAEDAALPGRATPIRVSGTHFVN
. * *.*
Mb0142c|M.bovis_AF2122/97 -------------------GCFWGLQDLIRNQPGVVSTRVGYSGGNIPNA
Rv0137c|M.tuberculosis_H37Rv -------------------GCFWGLQDLIRNQPGVVSTRVGYSGGNIPNA
MMAR_0347|M.marinum_M -------------------GCFWGMQDLIRKQPGVIATRVGYSGGDVANA
MUL_4773|M.ulcerans_Agy99 -------------------GCFWGMQDLIRKQPGVIATRVGYSGGDVANA
MAV_5159|M.avium_104 -------------------GCFWGMQELIRKQPGVVSTRVGYTGGDVPNA
MLBr_02647|M.leprae_Br4923 -------------------GCFWGIQNLFRRQPGVISTRVGYTGGGTPNA
Mflv_1105|M.gilvum_PYR-GCK -------------------GCFWGMQDLIRKQPGVVSTRVGYTGGQNDHP
Mvan_5709|M.vanbaalenii_PYR-1 -------------------GCFWGMQDLIRKQPGVVSTRVGYTGGQNDHP
TH_0081|M.thermoresistible__bu -------------------GCFWGMQELIRKQPGVVSTRVGYTGGENDHP
MSMEG_4417|M.smegmatis_MC2_155 -------------------GCFWGLEDLIRALPGVISTRVGYTGGDNDHP
MAB_0120|M.abscessus_ATCC_1997 GHTTVPPFPDGCELAVFGEGCFWGAERGFWKLPGVYSTAVGYAGGYTPNP
***** : : *** :* ***:** :.
Mb0142c|M.bovis_AF2122/97 TYRN----HGTHAEAVEIIFDPTVTDYRTLLEFFFQIHDPTTKDRQGNDR
Rv0137c|M.tuberculosis_H37Rv TYRN----HGTHAEAVEIIFDPTVTDYRTLLEFFFQIHDPTTKDRQGNDR
MMAR_0347|M.marinum_M TYRN----HGTHAEAVEIIFDPQATDYRTLLEFFFQIHDPTTPNRQGNDR
MUL_4773|M.ulcerans_Agy99 TYRN----HGTHAEAVEIIFDPQATDYRTLLEFFFQIHDPTTPNRQGNDR
MAV_5159|M.avium_104 TYRN----HGTHAEAVEIIYDPAVTDYRQILEFFFQIHDPTTKDRQGNDR
MLBr_02647|M.leprae_Br4923 TYRN----HGMHAEAVEIIYDPAITDYRTLLEFFFQIHDPTTRNRQGNDQ
Mflv_1105|M.gilvum_PYR-GCK TYRN----HPGHAEAIEIVYDPAQTDYRALLEFFFQIHDPTTKNRQGNDI
Mvan_5709|M.vanbaalenii_PYR-1 TYRN----HPGHAEAIEIVYDPAQTDYRALLEFFFQIHDPTTKNRQGNDV
TH_0081|M.thermoresistible__bu TYRN----HPGHAEAIEIVYDPAQTDYRTLLEFFFQIHDPTTKDRQGNDI
MSMEG_4417|M.smegmatis_MC2_155 TYRN----HPGHAEAVEITYDPQQTDYRALLEFFFQIHDPTTKNRQGNDV
MAB_0120|M.abscessus_ATCC_1997 TYEEVCSGRTGHAEVVLVVFDPREVTYGQLLVQFWESHDPTQGMRQGNDR
**.: : ***.: : :** . * :* *:: **** *****
Mb0142c|M.bovis_AF2122/97 GTSYRSAIFYFDEQQKRIALDTIADVEASGLWP--GKVVTEVSPAG----
Rv0137c|M.tuberculosis_H37Rv GTSYRSAIFYFDEQQKRIALDTIADVEASGLWP--GKVVTEVSPAG----
MMAR_0347|M.marinum_M GTSYRSAIYYLDDEQKRVALDTIADVEASGLWP--GKVVTEVSPAG----
MUL_4773|M.ulcerans_Agy99 GTSYRSAIYYLDDEQKRVALDTIADVEASGLWP--GKVVTEVSPAG----
MAV_5159|M.avium_104 GPSYRSAIFYTDDEQKRIALDTIADVEASGLWP--GKVVTEVSPAG----
MLBr_02647|M.leprae_Br4923 GTSYRSAIFYLDDEQKKVALDTIAAVEASSLWP--GKVVTEVSLAE----
Mflv_1105|M.gilvum_PYR-GCK GTSYRSEIFYVDEEQRQIALDTIADVDASGLWP--GKVVTEVSPAP----
Mvan_5709|M.vanbaalenii_PYR-1 GTSYRSEIFYVDDEQRRIALDTIADVDASGLWP--GKVVTDVSPAP----
TH_0081|M.thermoresistible__bu GSSYRSAIFYLDEEQKRIAEDTIADVDASGLWP--GKVVTEVTPAG----
MSMEG_4417|M.smegmatis_MC2_155 GSSYRSAIFYLDDDQKRVAFDIVAEIDASGRWP--GPVATEVTPAG----
MAB_0120|M.abscessus_ATCC_1997 GTQYRSVIYTTSEGQAVQAAESAARYEAALADAGYGAITTEIAPLNGPVG
*..*** *: .: * * : * :*: . * :.*:::
Mb0142c|M.bovis_AF2122/97 ----DFWEAEPEHQDYLQRYPNGYTCHFVRPGWRLPRRTAESALRASLSP
Rv0137c|M.tuberculosis_H37Rv ----DFWEAEPEHQDYLQRYPNGYTCHFVRPGWRLPRRTAESALRASLSP
MMAR_0347|M.marinum_M ----DFWEAEPEHQDYLQRYPSGYTCHFIRPGWKLPRRATAGQ-------
MUL_4773|M.ulcerans_Agy99 ----DFWEAEPEHQHYLQRYPSGYTCHFIRPGWKLPRRATAGQ-------
MAV_5159|M.avium_104 ----DFWEAEPEHQDYLQRYPNGYTCHYIRPGWKLPRRATSGS-------
MLBr_02647|M.leprae_Br4923 ----DFWEAEPEHQDYLQHYPNGYTCHFVRPNWKLPRRAPADAKADS---
Mflv_1105|M.gilvum_PYR-GCK ----DFWEAEPEHQDYLERYPTGYTCHFPRPGWKLPKRAEV---------
Mvan_5709|M.vanbaalenii_PYR-1 ----EFWEAEPEHQDYLERYPSGYTCHFPRPGWKLPKRAEV---------
TH_0081|M.thermoresistible__bu ----TFWEAEPEHQDYLQRFPNGYTCHYPRPNWKLTVRSGIRGR------
MSMEG_4417|M.smegmatis_MC2_155 ----VFWEAEPEHQDYLQRNPGGYTCHWVRPEWILDPEADPETASDSGSD
MAB_0120|M.abscessus_ATCC_1997 HASRTFFYAEEYHQQYLAKNPNGY-CGLGGTSVSCPIGLDVPPGGPAEA-
*: ** **.** : * ** * .
Mb0142c|M.bovis_AF2122/97 ELGT--------
Rv0137c|M.tuberculosis_H37Rv ELGT--------
MMAR_0347|M.marinum_M ------------
MUL_4773|M.ulcerans_Agy99 ------------
MAV_5159|M.avium_104 ------------
MLBr_02647|M.leprae_Br4923 ------------
Mflv_1105|M.gilvum_PYR-GCK ------------
Mvan_5709|M.vanbaalenii_PYR-1 ------------
TH_0081|M.thermoresistible__bu ------------
MSMEG_4417|M.smegmatis_MC2_155 VSAASATASHHD
MAB_0120|M.abscessus_ATCC_1997 ------------