For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAPKVLFIHNEHMCTEAMLGDAFSECGFDIETFEVVPPERVETPAGDVAFPDPTAYDVIVPLGARWPVYE QSLVGTWVTAEMDMMRKAADAGVGILGVCFGGQLLAQTFGGSVARAETAEVGWFELDTDDAGLIAPGPWF QWHFDRWTVPPGATEIARTSRSSQAFVLGRALALQFHPEVDVDLLEGWLADDREGISGKLGYNHDDLRLR TKELVDDAAVRVRELVRAFLDKVVRADPAS
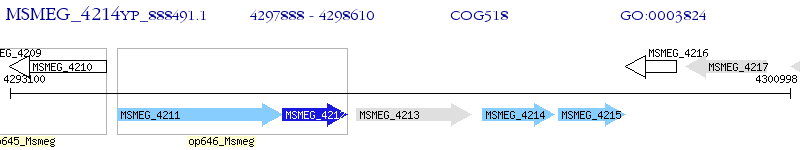
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4214 | - | - | 100% (240) | glutamine amidotransferase |
| M. smegmatis MC2 155 | MSMEG_1610 | guaA | 2e-06 | 33.66% (101) | bifunctional GMP synthase/glutamine amidotransferase protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3429c | guaA | 3e-07 | 34.34% (99) | GMP synthase |
| M. gilvum PYR-GCK | Mflv_3000 | - | 1e-88 | 65.24% (233) | glutamine amidotransferase class-I |
| M. tuberculosis H37Rv | Rv3396c | guaA | 3e-07 | 34.34% (99) | GMP synthase |
| M. leprae Br4923 | MLBr_00395 | guaA | 4e-09 | 33.33% (99) | GMP synthase |
| M. abscessus ATCC 19977 | MAB_3718c | - | 2e-06 | 35.35% (99) | GMP synthase |
| M. marinum M | MMAR_1148 | guaA | 2e-07 | 33.33% (99) | GMP synthase, GuaA |
| M. avium 104 | MAV_4348 | guaA | 9e-08 | 33.33% (99) | bifunctional GMP synthase/glutamine amidotransferase protein |
| M. thermoresistible (build 8) | TH_0484 | guaA | 5e-08 | 36.36% (99) | PROBABLE GMP SYNTHASE |
| M. ulcerans Agy99 | MUL_0913 | guaA | 1e-07 | 33.33% (99) | GMP synthase |
| M. vanbaalenii PYR-1 | Mvan_3511 | - | 6e-89 | 65.67% (233) | glutamine amidotransferase class-I |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00395|M.leprae_Br4923 ---------MADSAGLDQPEPSPRPVLVVDFGAQYAQLIARRVREARVFS
MAV_4348|M.avium_104 -------------------------MLVVDFGAQYAQLIARRVREARVFS
Mb3429c|M.bovis_AF2122/97 ---------MVQPADIDVPETPARPVLVVDFGAQYAQLIARRVREARVFS
Rv3396c|M.tuberculosis_H37Rv ---------MVQPADIDVPETPARPVLVVDFGAQYAQLIARRVREARVFS
MMAR_1148|M.marinum_M ---------MAEPVDLDVAGPVGRPVLVVDFGAQYAQLIARRVREARVFS
MUL_0913|M.ulcerans_Agy99 -------MPVAEPVDLDVAGPVGRPVLVVDFGAQYAQLIARRVREARVFS
TH_0484|M.thermoresistible__bu ---------VSSSADVN----TSRPVLVIDFGAQYAQLIARRIREARVYS
MAB_3718c|M.abscessus_ATCC_199 ---------MAQTEQHG-----DRPVLVIDFGAQYAQLIARRVREARVFS
Mflv_3000|M.gilvum_PYR-GCK MARVTSKVLFLLNEHIATEALLGDAFAERGFDIDTFEVVSAARAHSPAGE
Mvan_3511|M.vanbaalenii_PYR-1 MADVASKVLFLVNEHIATEALLGDAFAEQGFDVDTFEVVPAERVGAPAGE
MSMEG_4214|M.smegmatis_MC2_155 ---MAPKVLFIHNEHMCTEAMLGDAFSECGFDIETFEVVPPERVETPAGD
. .*. : :::. : . .
MLBr_00395|M.leprae_Br4923 EVIPHTTSIEEITARDPLAIVLSGGPASVYTEGAPQLDPALFDLGLPVFG
MAV_4348|M.avium_104 EVIPHTATIEEIKARQPRALVLSGGPSSVYEPGAPQLDPAVFDLGVPVFG
Mb3429c|M.bovis_AF2122/97 EVIPHTASIEEIRARQPVALVLSGGPASVYADGAPKLDPALLDLGVPVLG
Rv3396c|M.tuberculosis_H37Rv EVIPHTASIEEIRARQPVALVLSGGPASVYADGAPKLDPALLDLGVPVLG
MMAR_1148|M.marinum_M EVIPHTTSIEEIKARDPLALVLSGGPASVYAPGAPQLDPALFDLGLPVFG
MUL_0913|M.ulcerans_Agy99 EVIPHTTSIEEIKARDPLALVLSGGPASVYAPGAPQLDPALFDLGLPVFG
TH_0484|M.thermoresistible__bu EVVPHTATVEEIKAKDPQAIVLSGGPASVYAEDAPRLDPGLFDLGVPVFG
MAB_3718c|M.abscessus_ATCC_199 EVIPHSASVEEIKGRNPRAIVLSGGPSSVYEEGAPQLDPGVFDLDVPVFG
Mflv_3000|M.gilvum_PYR-GCK VTFPDPTRYDVIVPLGATWAVYDQTLLDTWVSTEIQMMRDAADAGVALLG
Mvan_3511|M.vanbaalenii_PYR-1 VTFPDPGGYDVIVPLGASWPVYDESLRDTWVGTEMQLLRDAADAGIAVLG
MSMEG_4214|M.smegmatis_MC2_155 VAFPDPTAYDVIVPLGARWPVYEQSLVGTWVTAEMDMMRKAADAGVGILG
..*.. : * . * . ..: : * .: ::*
MLBr_00395|M.leprae_Br4923 ICYGFQAMAQVLGGTVARTKTSEYGRTELKVLGGDLHSGLPGVQPVWMSH
MAV_4348|M.avium_104 ICYGFQAMAQALGGTVAHTGTSEYGRTELKVLGGELHSGLPGVQPVWMSH
Mb3429c|M.bovis_AF2122/97 ICYGFQAMAQALGGIVAHTGTREYGRTELKVLGGKLHSDLPEVQPVWMSH
Rv3396c|M.tuberculosis_H37Rv ICYGFQAMAQALGGIVAHTGTREYGRTELKVLGGKLHSDLPEVQPVWMSH
MMAR_1148|M.marinum_M ICYGFQVMAQALGGTVAHTGTSEYGRTELKVLGGDLHSDLPDIQPVWMSH
MUL_0913|M.ulcerans_Agy99 ICYGFQVMAQALGGTVAHTGTSEYGRTELKVLGGDLHSDLPDIQPVWMSH
TH_0484|M.thermoresistible__bu ICYGFQAMAQALGGTVTRTGTREYGRTELTVLGGQLHSDLPERQPVWMSH
MAB_3718c|M.abscessus_ATCC_199 ICYGFQAMAQVLGGTVAHTGTSEYGRTELTVAGGDLHEGLPDVQPVWMSH
Mflv_3000|M.gilvum_PYR-GCK VCFGGQLLAQAFGGTVARAPRPEIGWCEVASD----RPDLVPGGPWFQWH
Mvan_3511|M.vanbaalenii_PYR-1 VCFGGQLLAQAFGGSVARSPRPEIGWYDVLSD----RPEVVPGGPWFQWH
MSMEG_4214|M.smegmatis_MC2_155 VCFGGQLLAQTFGGSVARAETAEVGWFELDTD----DAGLIAPGPWFQWH
:*:* * :**.:** *::: * * :: : * : *
MLBr_00395|M.leprae_Br4923 GDAVTAAPDGFDVAASSSGAPVAAFENRDRRLAGVQYHPEVMHTPHGQQV
MAV_4348|M.avium_104 GDAVTAAPDGFEVVASSPGAVVAAFENRARRLAGVQYHPEVMHTPHGQQV
Mb3429c|M.bovis_AF2122/97 GDAVTAAPDGFDVVASSAGAPVAAFEAFDRRLAGVQYHPEVMHTPHGQQV
Rv3396c|M.tuberculosis_H37Rv GDAVTAAPDGFDVVASSAGAPVAAFEAFDRRLAGVQYHPEVMHTPHGQQV
MMAR_1148|M.marinum_M GDAVTAAPDGFEVVASSAGAAVAAFENRERRLAGVQYHPEVMHTPHGQQI
MUL_0913|M.ulcerans_Agy99 GDAVTAAPDGFEVVASSAGAAVAAFENRERRLAGVQYHPEVMHTPHGQQI
TH_0484|M.thermoresistible__bu ADAVTEAPAGFEVVATSPGSPVAAFEDPARRLAGVQYHPEVIHSPHGQQV
MAB_3718c|M.abscessus_ATCC_199 GDAVTEAPQGFTVVASSEGAPVAAFENRERRLAGVQYHPEVLHSPHGQQV
Mflv_3000|M.gilvum_PYR-GCK HDRWTLPPGATEIARTAHASQAFVLG----RTLALQFHPEVDTD------
Mvan_3511|M.vanbaalenii_PYR-1 FDRWTPPPGATEIARTPNASQAFVLG----RTLALQFHPEVDGD------
MSMEG_4214|M.smegmatis_MC2_155 FDRWTVPPGATEIARTSRSSQAFVLG----RALALQFHPEVDVD------
* * .* . :. :. .: . .: * .:*:****
MLBr_00395|M.leprae_Br4923 LSRFLHDFAGLDADWTAANIAGVLVEQVRAQIGNGHAICGLSGGVDSAVA
MAV_4348|M.avium_104 LSRFLHDFAGLGADWTAANIAGVLVEQVRAQIGDGHAICGLSGGVDSAVA
Mb3429c|M.bovis_AF2122/97 LSRFLHDFAGLGAQWTPANIANALIEQVRTQIGDGHAICGLSGGVDSAVA
Rv3396c|M.tuberculosis_H37Rv LSRFLHDFAGLGAQWTPANIANALIEQVRTQIGDGHAICGLSGGVDSAVA
MMAR_1148|M.marinum_M LGRFLHDFAGIGARWTPANIANALIEQVRTQIGDGHAICGLSGGVDSAVA
MUL_0913|M.ulcerans_Agy99 LGRFLHDFAGIGARWTPANIANALIEQVRTQIGDGHAICGLSGGVDSAVA
TH_0484|M.thermoresistible__bu LGRFLHEFAGIGASWTPANIAESLIEQVREQIGDGRAICGLSGGVDSAVA
MAB_3718c|M.abscessus_ATCC_199 LSRFLHEFAGIESAWTAANIAETLVEQVRTQIGEGRALCALSGGVDSAVA
Mflv_3000|M.gilvum_PYR-GCK ----------LLKIWLAT---------------DDHGDVDRAG-------
Mvan_3511|M.vanbaalenii_PYR-1 ----------LLKMWLAA---------------DRDGDVIAAG-------
MSMEG_4214|M.smegmatis_MC2_155 ----------LLEGWLAD---------------DREGISGKLG-------
: * . : . *
MLBr_00395|M.leprae_Br4923 AALVQRAIGDRLTCIFVDHGLLRDGERGQVQRDFVAATGAKLVTVDAAET
MAV_4348|M.avium_104 AALVQRAIGDRLTCVFVDHGLLRAGERAQVQRDFVAATGANLVTVDAADT
Mb3429c|M.bovis_AF2122/97 AALVQRAIGDRLTCVFVDHGLLRAGERAQVQRDFVAATGANLVTVDAAET
Rv3396c|M.tuberculosis_H37Rv AALVQRAIGDRLTCVFVDHGLLRAGERAQVQRDFVAATGANLVTVDAAET
MMAR_1148|M.marinum_M AALVQRAIGDRLTCVFVDHGLLRAGERAQVERDFVAATKANLVTVDVADT
MUL_0913|M.ulcerans_Agy99 AALVQRVIGDRLTCVFVDHGLLRAGERAQVERDFVAATKANLVTVDVADT
TH_0484|M.thermoresistible__bu AALVQRAIGDRLTCVFVDHGLLRAGERAQVQRDFVAATGAKLVTVDTSER
MAB_3718c|M.abscessus_ATCC_199 AALVQRAIGDRLTCVFVDHGLLRSGERAQVEHDFVAATGATLVTVDASET
Mflv_3000|M.gilvum_PYR-GCK --------------------------------------------LTHDEM
Mvan_3511|M.vanbaalenii_PYR-1 --------------------------------------------LDHDEL
MSMEG_4214|M.smegmatis_MC2_155 --------------------------------------------YNHDDL
:
MLBr_00395|M.leprae_Br4923 FLQALSGVTNPEGKRKIIGRQFIRAFEGAVRDLLGDSTFD--SGIEFLVQ
MAV_4348|M.avium_104 FLQALAGVTNPEGKRKIIGRQFIRAFEGAVRDVLGD------KPVEFLVQ
Mb3429c|M.bovis_AF2122/97 FLEALSGVSAPEGKRKIIGRQFIRAFEGAVRDVLDG------KTAEFLVQ
Rv3396c|M.tuberculosis_H37Rv FLEALSGVSAPEGKRKIIGRQFIRAFEGAVRDVLDG------KTAEFLVQ
MMAR_1148|M.marinum_M FLAALSGVTDPEGKRKIIGRQFIRAFEGAVRDVLDG------RDIEFLVQ
MUL_0913|M.ulcerans_Agy99 FLAALSGVTDPEGKRKIIGRQFIRAFEGAVRDVLDG------RDVEFLVQ
TH_0484|M.thermoresistible__bu FLEALSGVTNPEGKRKIIGREFIRAFEGAVREALGENAAANDDEVEFLVQ
MAB_3718c|M.abscessus_ATCC_199 FLGELAGVTDPEAKRKIIGREFIRSFEGAVSDVVGDSRRDDTAGIQFLVQ
Mflv_3000|M.gilvum_PYR-GCK LTRTEQLADDVASRIRVLVRGFLTQVAGRSLPS-----------------
Mvan_3511|M.vanbaalenii_PYR-1 LTRTTELAQDTGPRIRSLVRGFLTHVAGHSRPS-----------------
MSMEG_4214|M.smegmatis_MC2_155 RLRTKELVDDAAVRVRELVRAFLDKVVRADPAS-----------------
. : : : * *: .
MLBr_00395|M.leprae_Br4923 GTLYPDVVESGGGSGTANIKSHHNVGGLPDNLRFKLVEPLRLLFKDEVRA
MAV_4348|M.avium_104 GTLYPDVVESGRGSGTANIKSHHNVGGLPGDLKFTLVEPLRLLFKDEVRA
Mb3429c|M.bovis_AF2122/97 GTLYPDVVESGGGSGTANIKSHHNVGGLPDDLKFTLVEPLRLLFKDEVRA
Rv3396c|M.tuberculosis_H37Rv GTLYPDVVESGGGSGTANIKSHHNVGGLPDDLKFTLVEPLRLLFKDEVRA
MMAR_1148|M.marinum_M GTLYPDVVESGGGSGTANIKSHHNVGGLPDDLKFALVEPLRLLFKDEVRA
MUL_0913|M.ulcerans_Agy99 GTLYPDVVESGGGSGTANIKSHHNVGGLPDDLKFTLVEPLRLLFKDEVRA
TH_0484|M.thermoresistible__bu GTLYPDVVESGGGAGTANIKSHHNVGGLPDDLKFKLVEPLRLLFKDEVRA
MAB_3718c|M.abscessus_ATCC_199 GTLYPDVVESGGGSGTANIKSHHNVGGLPEDLTFTLVEPLRLLFKDEVRA
Mflv_3000|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3511|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4214|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00395|M.leprae_Br4923 VGRQLDLPEEIVARQPFPGPGLGIRIVGEVTAERLDTLRRADSIAREELT
MAV_4348|M.avium_104 VGRELGLPEEIVARQPFPGPGLGIRIVGEVTATRLDTLRRADSIAREELT
Mb3429c|M.bovis_AF2122/97 VGRELGLPEEIVARQPFPGPGLGIRIVGEVTAKRLDTLRHADSIVREELT
Rv3396c|M.tuberculosis_H37Rv VGRELGLPEEIVARQPFPGPGLGIRIVGEVTAKRLDTLRHADSIVREELT
MMAR_1148|M.marinum_M VGRELGLPEEIVARQPFPGPGLGIRIVGEVTGQRLDTLRRADSIAREELT
MUL_0913|M.ulcerans_Agy99 VGRELGLPEEIVARQPFPGPGLGIRIVGEVTGQRLDTLRRADSIARKELT
TH_0484|M.thermoresistible__bu VGRELGLPEEIVARQPFPGPGLAIRIVGEVTAERLETLRRADAIVREELT
MAB_3718c|M.abscessus_ATCC_199 VGRELGLPEEIVGRQPFPGPGLAIRIVGEVTPDRLDTLRRADAIAREELT
Mflv_3000|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3511|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4214|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00395|M.leprae_Br4923 TAGLDYQIWQCPVVLLADVRSVGVQGDNRSYGHPIVLRPVSSEDAMTADW
MAV_4348|M.avium_104 AAGLDSLIWQCPVVLLGEVRSVGVQGDNRTYGHPIVLRPVTSEDAMTADW
Mb3429c|M.bovis_AF2122/97 AAGLDNQIWQCPVVLLADVRSVGVQGDGRTYGHPIVLRPVSSEDAMTADW
Rv3396c|M.tuberculosis_H37Rv AAGLDNQIWQCPVVLLADVRSVGVQGDGRTYGHPIVLRPVSSEDAMTADW
MMAR_1148|M.marinum_M AAGLDNQIWQCPVVLLADVRSVGVQGDNRTYGHPIVLRPVSSEDAMTADW
MUL_0913|M.ulcerans_Agy99 AAGLDNQIWQCPVVLLADVRSVGVQGDNRTYGHPIVLRPVSSEDAMTADW
TH_0484|M.thermoresistible__bu AAGLDHQIWQCPVVLLADVRSVGVQGDGRTYGHPIVLRPVSSEDAMTADW
MAB_3718c|M.abscessus_ATCC_199 AAGQDRNIWQCPVVLLADVRSVGVQGDGRTYGHPIVLRPVSSEDAMTADW
Mflv_3000|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3511|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4214|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00395|M.leprae_Br4923 TWVPYEVLECISTRITNEVAEVNRVVLDITNKPPGTIEWE
MAV_4348|M.avium_104 TRVPYEVLERISTRITNEVPEVNRVVLDITSKPPGTIEWE
Mb3429c|M.bovis_AF2122/97 TRVPYEVLERISTRITNEVAEVNRVVLDITSKPPATIEWE
Rv3396c|M.tuberculosis_H37Rv TRVPYEVLERISTRITNEVAEVNRVVLDITSKPPATIEWE
MMAR_1148|M.marinum_M TRVPFEVLERISTRITNEVPEVNRVVLDVTSKPPGTIEWE
MUL_0913|M.ulcerans_Agy99 TRVPFEVLERISTRITNEVPEVNRVVLDVTSKPPGTIEWE
TH_0484|M.thermoresistible__bu TRVPYDVLERISTRITNEVAEVNRVVLDITSKPPATIEWE
MAB_3718c|M.abscessus_ATCC_199 TRLPYEVLERISTRITNEVPEVNRVVLDITSKPPGTIEWE
Mflv_3000|M.gilvum_PYR-GCK ----------------------------------------
Mvan_3511|M.vanbaalenii_PYR-1 ----------------------------------------
MSMEG_4214|M.smegmatis_MC2_155 ----------------------------------------