For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTTRDLTALFDPKSVAVLGASNDETKYGNWMSVQALRMTGTRPVYLINRRGDSVLGQPPLRSLAEVDGAV DLVAITVPAHSFEAAVEDALAAGARAIVGVTAGFAELGPEGRALQDRVVDRVRSAGAVLLGPNCLGVIDS TTNLTFASNPLPGGRVALLSQSGNMALEVSGFLQSRGHGFSRFASLGNQADLGVADLIDSCVAHNGTDLI ALYCEDFGDGRSFVSAAAAAAAANKPVLLLTVGGSEASVRGAQSHTGALTSDSAVIDAACHAAGIYRVTS PRQLADVAATLLTYGRRRVHRVAVIADGGGHAGVGSDVVEANGLSVPRFPTALSTALRPLLPPSAGVTNP VDIAGGGEQDIGCFASVLDAVMSSAEIDSVLLTGYFGGYAEYGEGLARDELDVAARIATIAQTHQKPLVV HTMHSASKAARVLIDAGVPVFAAVDDAAGALAVIDRGTDPRPLPSLPGTAEKPLSSTDYWDARELFRAAG VTFPEAKLVSDAAQARAAAESLGYPVVVKAMGLLHKSDVGGVALGLSDPHAVAEAVEGMRHRLNPPGFCV EAMADLNSGVEIIVGVQRDPRFGPVAMVGLGGVYTEVLADVAFALAPVDSGAARALLESLRASALFTGLR GRAPVDLDAAARAIAAITEVAVTHPEITELEINPLLLTPDHALGLDARIVLDNQPTH
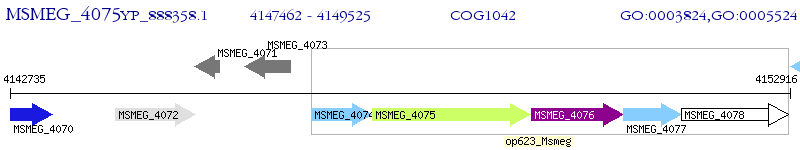
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4075 | - | - | 100% (687) | CoA-binding protein |
| M. smegmatis MC2 155 | MSMEG_0666 | - | 7e-64 | 29.06% (702) | acetyl-CoA synthetase, putative |
| M. smegmatis MC2 155 | MSMEG_5524 | sucD | 6e-07 | 28.17% (252) | succinyl-CoA synthetase subunit alpha |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0976 | sucC | 6e-08 | 30.04% (223) | succinyl-CoA synthetase subunit beta |
| M. gilvum PYR-GCK | Mflv_1866 | - | 7e-08 | 28.46% (246) | succinyl-CoA synthetase subunit alpha |
| M. tuberculosis H37Rv | Rv0951 | sucC | 6e-08 | 30.04% (223) | succinyl-CoA synthetase subunit beta |
| M. leprae Br4923 | MLBr_00156 | sucD | 2e-06 | 24.90% (253) | succinyl-CoA synthetase subunit alpha |
| M. abscessus ATCC 19977 | MAB_1056 | sucC | 4e-09 | 29.65% (226) | succinyl-CoA synthetase subunit beta |
| M. marinum M | MMAR_4549 | sucD | 2e-07 | 26.46% (257) | succinyl-CoA synthetase (alpha chain) SucD |
| M. avium 104 | MAV_1075 | - | 1e-06 | 26.41% (284) | succinyl-CoA synthetase subunit alpha |
| M. thermoresistible (build 8) | TH_1772 | sucD | 2e-06 | 25.54% (231) | PROBABLE SUCCINYL-CoA SYNTHETASE (ALPHA CHAIN) SUCD |
| M. ulcerans Agy99 | MUL_4719 | sucD | 6e-07 | 25.69% (253) | succinyl-CoA synthetase subunit alpha |
| M. vanbaalenii PYR-1 | Mvan_4870 | sucC | 8e-09 | 30.63% (222) | succinyl-CoA synthetase subunit beta |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_1075|M.avium_104 ------MSIFLNKDSKVIVQG--ITGGE-----------GTKHTALMLKA
TH_1772|M.thermoresistible__bu ------MSIFLNKNSKVIVQG--ITGGE-----------GTKHTALMLKA
MMAR_4549|M.marinum_M ------MSIFLNKDNKVIVQG--ITGSE-----------ATVHTARMLKA
MUL_4719|M.ulcerans_Agy99 ------MSIFLNKDNKVIVQG--ITGSE-----------ATVHTARMLKA
MLBr_00156|M.leprae_Br4923 ------MAIFLTKDNTVIVQG--ITGSE-----------AAVHTARMLKA
Mflv_1866|M.gilvum_PYR-GCK ------MSIFLNADSKVIVQG--ITGGE-----------GTKHTKLMLKA
Mb0976|M.bovis_AF2122/97 ------MDLFEYQAKELFAKHNVPSTPG-----------RVTDTAEGAKA
Rv0951|M.tuberculosis_H37Rv ------MDLFEYQAKELFAKHNVPSTPG-----------RVTDTAEGAKA
Mvan_4870|M.vanbaalenii_PYR-1 ------MDLFEYQAKELFAKHNVPTTPG-----------RVTESAEDAKA
MAB_1056|M.abscessus_ATCC_1997 ------MDLFEYQAKELFAKHNVPTTPG-----------RVTTTAEDAKA
MSMEG_4075|M.smegmatis_MC2_155 MTTRDLTALFDPKSVAVLGASNDETKYGNWMSVQALRMTGTRPVYLINRR
:* :: : . :
MAV_1075|M.avium_104 GTQVVGGVNARKAGTTVSHVDPVG-KDVELPVFGTVSEAMKETGANVS--
TH_1772|M.thermoresistible__bu GTQIVGGVNARKAGTTVSHTDKDG-KDVELPVFGTVEEAMKETGADVS--
MMAR_4549|M.marinum_M GTQIVGGVNARKAGTTVTHEDKGG-RVIKLPVFGGVAEAMEKTGADVS--
MUL_4719|M.ulcerans_Agy99 GTQIVGGVNARKAGTTVTHEDKGG-RVIKLPVFGGVAEAMEKTGADVS--
MLBr_00156|M.leprae_Br4923 GTQIVGGVNARKAGTNVTHEDKHG-RIITLPVFGTVAEAIENTGADVS--
Mflv_1866|M.gilvum_PYR-GCK GTQIVGGVNARKAGTEVTHQDKDG-KDVNLPVFGSVAEAMKETGADVS--
Mb0976|M.bovis_AF2122/97 IATEIGRPVMVKAQVKIGGRGKAGGVKYAATPQDAYEHAKNILGLDIK--
Rv0951|M.tuberculosis_H37Rv IATEIGRPVMVKAQVKIGGRGKAGGVKYAATPQDAYEHAKNILGLDIK--
Mvan_4870|M.vanbaalenii_PYR-1 IAEEIGKPVMVKAQVKVGGRGKAGGVKYAATPEDALTHAQNILGLDIK--
MAB_1056|M.abscessus_ATCC_1997 IAEEIGKPVMIKAQVKVGGRGKAGGVKYAATPDDALTHAQNILGLDIK--
MSMEG_4075|M.smegmatis_MC2_155 GDSVLGQPPLRSLAEVDGAVDLVAITVPAHSFEAAVEDALAAGARAIVGV
:* . . . . .* . :
MAV_1075|M.avium_104 --------------VVFVPPKFAKDA-----------IIEAIDAEIPLLV
TH_1772|M.thermoresistible__bu --------------VVFVPPKFAKDA-----------IIEAIDAEIALLV
MMAR_4549|M.marinum_M --------------IIFVPPKFAKDA-----------IIEAIDAEIPLLV
MUL_4719|M.ulcerans_Agy99 --------------IIFVPPKFAKDA-----------IIEAIDAEIPLLV
MLBr_00156|M.leprae_Br4923 --------------IIFVPPRFAKEA-----------IIEAVEAEIPLLV
Mflv_1866|M.gilvum_PYR-GCK --------------IAFVPPAFSKDA-----------IIEAIDAEIPLLV
Mb0976|M.bovis_AF2122/97 --------------GHIVKKLLVAEASDIAEEYYLSFLLDRANRTYLAMC
Rv0951|M.tuberculosis_H37Rv --------------GHIVKKLLVAEASDIAEEYYLSFLLDRANRTYLAMC
Mvan_4870|M.vanbaalenii_PYR-1 --------------GHIVKKLLVAEASDIAEEYYISFLLDRANRTYLAMC
MAB_1056|M.abscessus_ATCC_1997 --------------GHIVKKILVAEASDIAEEYYISFLLDRANRTYLAMC
MSMEG_4075|M.smegmatis_MC2_155 TAGFAELGPEGRALQDRVVDRVRSAGAVLLGPNCLGVIDSTTNLTFASNP
* . . : . :
MAV_1075|M.avium_104 VITEGIPVQDSAYAWAYNVDKGQKTRIIGPN-------------------
TH_1772|M.thermoresistible__bu VITEGIPVQDTAYAWAYNVKRGQKTRIIGPN-------------------
MMAR_4549|M.marinum_M VITEGIPVQDTAYAWAYNLGKGGKTRIIGPN-------------------
MUL_4719|M.ulcerans_Agy99 VITEVIPVQDTAYAWAYNLGKGGKTRIIGPN-------------------
MLBr_00156|M.leprae_Br4923 IITEGIPVQDSVYAWAYNLNKGTKTRIIGPN-------------------
Mflv_1866|M.gilvum_PYR-GCK VITEGIPVQDTAYAWAYNVEKGEKTRIIGPN-------------------
Mb0976|M.bovis_AF2122/97 SVEGGMEIEEVAATKPERLAKVPVNAVKGVDLDFARSIAEQGHLPAEVLD
Rv0951|M.tuberculosis_H37Rv SVEGGMEIEEVAATKPERLAKVPVNAVKGVDLDFARSIAEQGHLPAEVLD
Mvan_4870|M.vanbaalenii_PYR-1 SVEGGMEIEEVAATKPERLAKVPVDAVKGVDEAFAREIAEKGHLPAEVLD
MAB_1056|M.abscessus_ATCC_1997 SVEGGVEIEVTAEENPDALAKVPVDAVKGVDLALAREIAEKGKLPAEVLD
MSMEG_4075|M.smegmatis_MC2_155 LPGGRVALLSQSGNMALEVSGFLQSRGHGFSRFASLGNQADLGVADLIDS
: : . : * .
MAV_1075|M.avium_104 -----------------------CPGIITPGEALAGITP-------ANIS
TH_1772|M.thermoresistible__bu -----------------------CPGIITPGEALAGITP-------ANIT
MMAR_4549|M.marinum_M -----------------------CPGIITPGQALVGITP-------ANIT
MUL_4719|M.ulcerans_Agy99 -----------------------CPGIITPGQALVGITP-------ANIT
MLBr_00156|M.leprae_Br4923 -----------------------CPGIITPGQSLVGITP-------ATIT
Mflv_1866|M.gilvum_PYR-GCK -----------------------CPGIITPGESLVGITP-------NNIT
Mb0976|M.bovis_AF2122/97 TAAVTIAKLWELFVAEDATLVEVNPLVRTPDHKILALDAKITLDGNADFR
Rv0951|M.tuberculosis_H37Rv TAAVTIAKLWELFVAEDATLVEVNPLVRTPDHKILALDAKITLDGNADFR
Mvan_4870|M.vanbaalenii_PYR-1 AAAVTIAKLWEVFVKEDATLVEVNPLVRTPDDQILALDGKVTLDANADFR
MAB_1056|M.abscessus_ATCC_1997 SAAVTIQKLWEVFVGEDATLVEVNPLVRTPDNQILALDGKVTLDGNADFR
MSMEG_4075|M.smegmatis_MC2_155 CVAHNGTDLIALYCEDFGDGRSFVSAAAAAAAANKPVLLLTVGGSEASVR
. :. : .
MAV_1075|M.avium_104 GPG---------------------------PVGLVSKSGTLTYQMMYELR
TH_1772|M.thermoresistible__bu GPG---------------------------PIGLVSKSGTLTYQMMFELS
MMAR_4549|M.marinum_M GAG---------------------------PVGLVSKSGTLTYQMMFELR
MUL_4719|M.ulcerans_Agy99 GAG---------------------------PVGLVSKSGTLTYQMMFELR
MLBr_00156|M.leprae_Br4923 GAG---------------------------PIGLVSKSGTLTYQMMFELS
Mflv_1866|M.gilvum_PYR-GCK GKG---------------------------PIGLVSKSGTLTYQMMYELR
Mb0976|M.bovis_AF2122/97 QPG---------------------------HAEFEDRAATDPLELKAKEH
Rv0951|M.tuberculosis_H37Rv QPG---------------------------HAEFEDRAATDPLELKAKEH
Mvan_4870|M.vanbaalenii_PYR-1 QPG---------------------------HAEFEDKDATDPLELKAKEN
MAB_1056|M.abscessus_ATCC_1997 QPG---------------------------HAEFEDKDATDPLELKAKEH
MSMEG_4075|M.smegmatis_MC2_155 GAQSHTGALTSDSAVIDAACHAAGIYRVTSPRQLADVAATLLTYGRRRVH
: . .* .
MAV_1075|M.avium_104 DFGFSTSIG----IGGDPIIG---------TTHIDAIEAFEKDPDTKIIV
TH_1772|M.thermoresistible__bu DFGFSTAIG----IGGDPIIG---------TTHIDAIEAFEKDPDTKLIV
MMAR_4549|M.marinum_M DFGFTTSIG----IGGDPVIG---------TTHIDAIEAFEKDPDTKLIV
MUL_4719|M.ulcerans_Agy99 DFGFTTSIG----IGGDPVIG---------TTHIDAIEAFEKDPDTKLIV
MLBr_00156|M.leprae_Br4923 DFGFSTAIG----IGGDPVIG---------TTHIDAIEAFEQDPDTKIIV
Mflv_1866|M.gilvum_PYR-GCK DLGFSTAIG----IGGDPVIG---------TTHIDAIEAFEKDPETKLIV
Mb0976|M.bovis_AF2122/97 DLNYVKLDGQVGIIGNGAGLA---------MSTLDVVAYAGEKHGGVKPA
Rv0951|M.tuberculosis_H37Rv DLNYVKLDGQVGIIGNGAGLV---------MSTLDVVAYAGEKHGGVKPA
Mvan_4870|M.vanbaalenii_PYR-1 DLNYVKLDGAVGIIGNGAGLV---------MSTLDVVAYAGEKHGGVKPA
MAB_1056|M.abscessus_ATCC_1997 DLNYVKLDGQVGIIGNGAGLV---------MSTLDVVAYAGENHNGVKPA
MSMEG_4075|M.smegmatis_MC2_155 RVAVIADGGGHAGVGSDVVEANGLSVPRFPTALSTALRPLLPPSAGVTNP
. * :*.. : .:
MAV_1075|M.avium_104 MIGEIGG----DAEERAADYIKAH--VSKPVVG-YVAGFTAPEGKTMGHA
TH_1772|M.thermoresistible__bu MIGEIGG----DAEERAAEYIKAN--VSKPVVG-YVAGFTAPEGKTMGHA
MMAR_4549|M.marinum_M MIGEIGG----DAEERAADFIKAN--VSKPVVG-YVAGFTAPEGKTMGHA
MUL_4719|M.ulcerans_Agy99 MIGEIGG----DAEERAADFIKAN--VSKPVVG-YVAGFTAPEGKTMGHA
MLBr_00156|M.leprae_Br4923 MIGEIGG----DAEERAADYIKAN--VSKPVVG-YVAGFTAPEGKTMGHA
Mflv_1866|M.gilvum_PYR-GCK MIGEIGG----DAEEKAADYIKAN--VTKPVVG-YVAGFTAPEGKTMGHA
Mb0976|M.bovis_AF2122/97 NFLDIGGGASAEVMAAGLDVVLGDQQVKSVFVN-VFGGITSCDAVATGIV
Rv0951|M.tuberculosis_H37Rv NFLDIGGGASAEVMAAGLDVVLGDQQVKSVFVN-VFGGITSCDAVATGIV
Mvan_4870|M.vanbaalenii_PYR-1 NFLDIGGGASAEVMAAGLDVILNDSQVKSVFVN-VFGGITACDAVANGIV
MAB_1056|M.abscessus_ATCC_1997 NFLDIGGGASAEVMAAGLDVILGDSQVKSVFVN-VFGGITACDAVANGIV
MSMEG_4075|M.smegmatis_MC2_155 VDIAGGGEQDIGCFASVLDAVMSSAEIDSVLLTGYFGGYAEYGEGLARDE
** : : : . .: ..* :
MAV_1075|M.avium_104 GAIVSGSSGTAAAKKEALEAAGVKVG---------------KTPSETAAL
TH_1772|M.thermoresistible__bu GAIVSGSSGTAQAKKEALEAAGVKVG---------------KTPSETARL
MMAR_4549|M.marinum_M GAIVSGSSGTAAAKKEALEAAGVQVG---------------KTPSETAML
MUL_4719|M.ulcerans_Agy99 GAIVSGSSGTAAAKKEALEAAGVQVG---------------KTPSETAVL
MLBr_00156|M.leprae_Br4923 GAIVSGSSGTAAVKKDALEAAGVKVG---------------KTPSETAVL
Mflv_1866|M.gilvum_PYR-GCK GAIVSDGAGTAAGKQEALEAAGVKVG---------------KTPSETAAK
Mb0976|M.bovis_AF2122/97 KALGMLGDEANKPLVVRLDGNNVEEGRRILTEANHPLVTLVATMDEAADK
Rv0951|M.tuberculosis_H37Rv KALGMLGDEANKPLVVRLDGNNVEEGRRILTEANHPLVTLVATMDEAADK
Mvan_4870|M.vanbaalenii_PYR-1 KALEILGDEANKPLVVRLDGNNVEEGRRILSEANHPLVIQADTMDSGADK
MAB_1056|M.abscessus_ATCC_1997 GALKTLGNTASKPLVVRLDGNKVEEGRAILAEFNHPLVIQAETMDAGADK
MSMEG_4075|M.smegmatis_MC2_155 LDVAARIATIAQTHQKPLVVHTMHSASKAARVLIDAGVPVFAAVDDAAGA
: * :. . : . *
MAV_1075|M.avium_104 AREILQSL------------------------------------------
TH_1772|M.thermoresistible__bu AREILQSL------------------------------------------
MMAR_4549|M.marinum_M AREILKGL------------------------------------------
MUL_4719|M.ulcerans_Agy99 AREILKGL------------------------------------------
MLBr_00156|M.leprae_Br4923 VREILCTL------------------------------------------
Mflv_1866|M.gilvum_PYR-GCK ARELLQNL------------------------------------------
Mb0976|M.bovis_AF2122/97 AAELASA-------------------------------------------
Rv0951|M.tuberculosis_H37Rv AAELASA-------------------------------------------
Mvan_4870|M.vanbaalenii_PYR-1 AAELANK-------------------------------------------
MAB_1056|M.abscessus_ATCC_1997 AAALAAASN-----------------------------------------
MSMEG_4075|M.smegmatis_MC2_155 LAVIDRGTDPRPLPSLPGTAEKPLSSTDYWDARELFRAAGVTFPEAKLVS
:
MAV_1075|M.avium_104 --------------------------------------------------
TH_1772|M.thermoresistible__bu --------------------------------------------------
MMAR_4549|M.marinum_M --------------------------------------------------
MUL_4719|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00156|M.leprae_Br4923 --------------------------------------------------
Mflv_1866|M.gilvum_PYR-GCK --------------------------------------------------
Mb0976|M.bovis_AF2122/97 --------------------------------------------------
Rv0951|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_4870|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_1056|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_4075|M.smegmatis_MC2_155 DAAQARAAAESLGYPVVVKAMGLLHKSDVGGVALGLSDPHAVAEAVEGMR
MAV_1075|M.avium_104 --------------------------------------------------
TH_1772|M.thermoresistible__bu --------------------------------------------------
MMAR_4549|M.marinum_M --------------------------------------------------
MUL_4719|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00156|M.leprae_Br4923 --------------------------------------------------
Mflv_1866|M.gilvum_PYR-GCK --------------------------------------------------
Mb0976|M.bovis_AF2122/97 --------------------------------------------------
Rv0951|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_4870|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_1056|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_4075|M.smegmatis_MC2_155 HRLNPPGFCVEAMADLNSGVEIIVGVQRDPRFGPVAMVGLGGVYTEVLAD
MAV_1075|M.avium_104 --------------------------------------------------
TH_1772|M.thermoresistible__bu --------------------------------------------------
MMAR_4549|M.marinum_M --------------------------------------------------
MUL_4719|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00156|M.leprae_Br4923 --------------------------------------------------
Mflv_1866|M.gilvum_PYR-GCK --------------------------------------------------
Mb0976|M.bovis_AF2122/97 --------------------------------------------------
Rv0951|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_4870|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_1056|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_4075|M.smegmatis_MC2_155 VAFALAPVDSGAARALLESLRASALFTGLRGRAPVDLDAAARAIAAITEV
MAV_1075|M.avium_104 -------------------------------------
TH_1772|M.thermoresistible__bu -------------------------------------
MMAR_4549|M.marinum_M -------------------------------------
MUL_4719|M.ulcerans_Agy99 -------------------------------------
MLBr_00156|M.leprae_Br4923 -------------------------------------
Mflv_1866|M.gilvum_PYR-GCK -------------------------------------
Mb0976|M.bovis_AF2122/97 -------------------------------------
Rv0951|M.tuberculosis_H37Rv -------------------------------------
Mvan_4870|M.vanbaalenii_PYR-1 -------------------------------------
MAB_1056|M.abscessus_ATCC_1997 -------------------------------------
MSMEG_4075|M.smegmatis_MC2_155 AVTHPEITELEINPLLLTPDHALGLDARIVLDNQPTH