For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MIGHSTMTHSKTKDHAAQADSDIAQRLSALGDDGRDLLFRRARTANTFSAEPVADALLEEVWDLAQWPPT SANGNPLRIVFIRSPEAKARLLPCIDEGNRPKVAAAPVTAVLATDTAFFESIPKLLPFKPEMRDLLAGRS DRDHIGEFNATLQIGYFIMAVRAVGLAAGPMAGFDRAGVDAEFLTGTTWRSLLTVNIGHPGPAAWYDRLP RHDPSDIVRFV
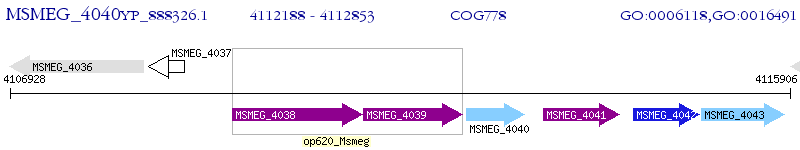
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4040 | - | - | 100% (221) | hypothetical protein MSMEG_4040 |
| M. smegmatis MC2 155 | MSMEG_6627 | - | 4e-05 | 27.43% (175) | nitroreductase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00758 | - | 3e-05 | 25.27% (186) | F420-0--gamma-glutamyl ligase |
| M. abscessus ATCC 19977 | MAB_4354 | - | 2e-54 | 52.31% (195) | hypothetical protein MAB_4354 |
| M. marinum M | MMAR_4721 | - | 6e-06 | 30.63% (111) | nitroreductase |
| M. avium 104 | MAV_0933 | - | 2e-06 | 29.93% (147) | nitroreductase family protein |
| M. thermoresistible (build 8) | TH_4633 | - | 2e-06 | 26.23% (183) | PUTATIVE putative nitroreductase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4040|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4354|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_4721|M.marinum_M --------------------------------------------------
MAV_0933|M.avium_104 --------------------------------------------------
TH_4633|M.thermoresistible__bu --------------------------------------------------
MLBr_00758|M.leprae_Br4923 MTSSDSHRSAPSPEHGTASTIEILPVAGLPEFRPGDDLSAALAAAAPWLR
MSMEG_4040|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4354|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_4721|M.marinum_M --------------------------------------------------
MAV_0933|M.avium_104 --------------------------------------------------
TH_4633|M.thermoresistible__bu --------------------------------------------------
MLBr_00758|M.leprae_Br4923 DGDVVVVTSKVVSKCEGRLVPAPEDTRGRNELRRKLINDETIRVLARKGR
MSMEG_4040|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4354|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_4721|M.marinum_M --------------------------------------------------
MAV_0933|M.avium_104 --------------------------------------------------
TH_4633|M.thermoresistible__bu --------------------------------------------------
MLBr_00758|M.leprae_Br4923 TLIIENGLGLVQAAAGVDGSNVGRGELALLPVNPDASAAVLRIGLRAMLG
MSMEG_4040|M.smegmatis_MC2_155 -----------------------------------------MIGHSTMTH
MAB_4354|M.abscessus_ATCC_1997 -----------------------------------------------MTD
MMAR_4721|M.marinum_M --------------------------------------------------
MAV_0933|M.avium_104 --------------------------------------------------
TH_4633|M.thermoresistible__bu --------------------------------------------------
MLBr_00758|M.leprae_Br4923 VNVAVVITDTMGRAWRNGQTDVAIGAAGLAVLHNYSGAVDRYGNELVVTE
MSMEG_4040|M.smegmatis_MC2_155 SKTKDHAAQADSDIAQRLSALGDDGRDLLFRR------------------
MAB_4354|M.abscessus_ATCC_1997 SN--GHIAVAS------LPVLDEAGRATLFTH------------------
MMAR_4721|M.marinum_M -----------------------------MST------------------
MAV_0933|M.avium_104 ----------------------------MMST------------------
TH_4633|M.thermoresistible__bu --VDRHAAADD-------PAMTSPDLWTVLRT------------------
MLBr_00758|M.leprae_Br4923 IAVADEVAAATDLVKGKLTAMPVAVVRGLSPTDDGSTAQHLLRNGPDDLF
MSMEG_4040|M.smegmatis_MC2_155 ------------------ARTANTFSAEPVADALLEEVWDLAQWPPTSAN
MAB_4354|M.abscessus_ATCC_1997 ------------------ARTANSFADTPIAREKLLEAWELARWAPTSVN
MMAR_4721|M.marinum_M ------------------ARTVRRFTDRPVDDATLTRCLEAATWAPSGAN
MAV_0933|M.avium_104 ------------------ARTIRRFTDEPVDDATLARCLGAARWAPSGAN
TH_4633|M.thermoresistible__bu ------------------ASAVRRYRDEPVDDVVVERCLRAASWAPSGGN
MLBr_00758|M.leprae_Br4923 WLGTTEALELGRQQAQLLRRSVRQFSDEPIAAELIETAVAEALTAPAPHH
: . : *: : * .*: :
MSMEG_4040|M.smegmatis_MC2_155 GNPLRIVFIRSPEAKARLLPCIDEGNRPKVAAA----PVTAVLATDTAFF
MAB_4354|M.abscessus_ATCC_1997 TQPLRILFVDKGEARDRLIPHMFDPNKAKTSTA----PAVAILATDTRFH
MMAR_4721|M.marinum_M AQGWRFVVLRSPEQRAVVAAAAARALAVIEPVYGMRRPAADDDSRRARTY
MAV_0933|M.avium_104 AQGWRFVVLRSPEQRAVVAKAAAQALEVIEPVYGMSRPAPDDHSRRARTY
TH_4633|M.thermoresistible__bu QQPWRFVVLRSPEVRAVITAAARRTWDVMADFYGFDKPGTADDPK-SRVL
MLBr_00758|M.leprae_Br4923 TRPVRFVWLQTPAVRTRLLDRMADKWRLDLASDALPADAIAQRVARGQIL
. *:: : . : :
MSMEG_4040|M.smegmatis_MC2_155 ESIPKLLPFKP-EMRDLLAGRSD-RDHIGEFN---ATLQIGYFIMAVRAV
MAB_4354|M.abscessus_ATCC_1997 DQIPTVFPLMP-EMAELFEANSELRSSTGIFN---AALQSAYFIMAIRAV
MMAR_4721|M.marinum_M RATYELHDRAG-EFTSVLFAQQHYPTASELLLGGSIFPAMQNFLLAARAQ
MAV_0933|M.avium_104 RATYELHDRAG-EFTSVLFAQAHYPTASELLLGGSIFPAMQNFLLAARAQ
TH_4633|M.thermoresistible__bu GAMYEHMQVGGGAPVCVLFCLRPQRGASDLQQGGSIFPAVQNFLLAARAQ
MLBr_00758|M.leprae_Br4923 YDAPEVIIPFMVPDGAHAYPDAARASAEHTMFIVAVGAAVQALLVALAVR
:::* .
MSMEG_4040|M.smegmatis_MC2_155 GLAAGPMAGFDRAGVDAEFLTGT----TWRSLLTVNIGHP-GPAAWYDRL
MAB_4354|M.abscessus_ATCC_1997 GLAAGPMGGFDADGVDREFFPDN----DWSTVLVVNIGHP-GENPWFGRL
MMAR_4721|M.marinum_M GLGA-CMTSWASYGGERLLREAVGVPEDWLLAGHIVIGWPQGKHGPLRRR
MAV_0933|M.avium_104 GLGA-CLTSWASYGGEQLLREAVGVPEGWMIAGHIVVGWPKGKHGPLRRR
TH_4633|M.thermoresistible__bu GLGA-AITLWHD-ACESELRSLVGIPDDWRIATLVTAGWPRGGHHPVRRK
MLBr_00758|M.leprae_Br4923 GLGS--CWIGSTIFADDLVRAELELPADWEPLGAIAIGYAHEPTDLREPV
**.: : . * : * .
MSMEG_4040|M.smegmatis_MC2_155 PRHDPSDIVRFV----------
MAB_4354|M.abscessus_ATCC_1997 PRLDPADVVRWA----------
MMAR_4721|M.marinum_M PLSDVVNLDRWDRPAFPGLGS-
MAV_0933|M.avium_104 PLAEFVNLDRWDAPADGLLSGL
TH_4633|M.thermoresistible__bu PVCEVAAVDRWDQAWVSRMR--
MLBr_00758|M.leprae_Br4923 RVADLLLRK-------------
: