For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
HSEVVTESDLELLHGLQIAPRISWADAARILRTTPATLAARWARLRREGLAWITAHRGGNPAPVMALIEV DCVPGARADVVTAICADRRAVTVEESTRGRDLLVTALTRDLEGLTEFVLDDLSSQPGVQTQRTSLITTLH RHGGDWRLGALDRRQESAFEAAAAAMRPSRSVRPPEDDGPLREVLAADGRASAADCARVTGRNPATVRRQ LARLLASGTVAFRCEVAQLVSGHPISSTWLVSVSPADQERTVAALSTLSELRMCASITGDATIAFTVWTS SLADITRLERRIGERLPWLSIRDSGVNLRTRKRMGWMLDPTGRATGVVVTPV*
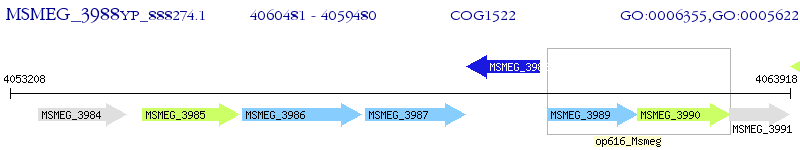
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3988 | - | - | 100% (333) | transcriptional regulator, AsnC family protein |
| M. smegmatis MC2 155 | MSMEG_1185 | - | 3e-52 | 36.06% (330) | transcriptional regulator, AsnC family protein |
| M. smegmatis MC2 155 | MSMEG_0838 | - | 5e-11 | 27.38% (336) | AsnC-family protein transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3319c | - | 0.0001 | 25.45% (110) | AsnC family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0397 | - | 7e-06 | 22.96% (135) | AsnC family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3291c | - | 3e-05 | 26.36% (110) | AsnC family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3380 | - | 2e-16 | 27.30% (315) | AsnC-family protein transcriptional regulator |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3319c|M.bovis_AF2122/97 --------------------------------------------------
Rv3291c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_0397|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_3988|M.smegmatis_MC2_155 MHSEVVTESDLELLHGLQIAPRISWADAARILRTTPATLAARWARLRREG
MAV_3380|M.avium_104 -----MEALDGHILHALQLSPRAPFRRIAEVVGAGEQTVARRYRALRRDG
Mb3319c|M.bovis_AF2122/97 --------------------------------------------------
Rv3291c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_0397|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_3988|M.smegmatis_MC2_155 LAWITAHRGGNPAPVMALIEVDCVPGARADVVTAICADRRAVTVEESTRG
MAV_3380|M.avium_104 VLRAVGVVNPRVYGECQWIVRVRAKPDDLLRLAEALVRRADVTHANVLSG
Mb3319c|M.bovis_AF2122/97 --------------------------------------------------
Rv3291c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_0397|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_3988|M.smegmatis_MC2_155 RDLLVTALTRDLEGLTEFVLDDLSSQPGVQTQRTSLITTLH---RHGGDW
MAV_3380|M.avium_104 GTELVCVVRAPLDDTEAGLLHRLPRTSTVLDMRVDLVINVFGAPMHAHWT
Mb3319c|M.bovis_AF2122/97 -----------------------MNEALDDIDRILVRELAADGRVTLSEL
Rv3291c|M.tuberculosis_H37Rv -----------------------MNEALDDIDRILVRELAADGRATLSEL
Mflv_0397|M.gilvum_PYR-GCK -----------MFDAIIGGMGDDDRIALDRTDEAILKALQDDGRISTADL
MSMEG_3988|M.smegmatis_MC2_155 RLGALDRRQESAFEAAAAAMRPSRSVRPPEDDGPLREVLAADGRASAADC
MAV_3380|M.avium_104 GHGHALDGHQVAALRPAHCAHPDRPVGPTPEDQPLLDALAEDGRISDSAL
* : * *** : :
Mb3319c|M.bovis_AF2122/97 ATRAGLSVSAVQSRVRRLESRGVVQGYSARINPEAVGHLLSAFVAITPLD
Rv3291c|M.tuberculosis_H37Rv ATRAGLSVSAVQSRVRRLESRGVVQGYSARINPEAVGHLLSAFVAITPLD
Mflv_0397|M.gilvum_PYR-GCK ARAVSLSPSSTADRVRRLVDAGVITGYTATVSATALGYTITAFVRLAYPS
MSMEG_3988|M.smegmatis_MC2_155 ARVTGRNPATVRRQLARLLASGTVA-FRCEVAQLVSGHPISS-TWLVSVS
MAV_3380|M.avium_104 AGLTGWSPARVKRRMVALQGSGTLS-FDLDVLPERLGFMLHAIIWISVVP
* .. . : . :: * *.: : : *. : : :
Mb3319c|M.bovis_AF2122/97 PSQPDDAPARLEHIEEVESCYSVAGEESYVLLVRVASARALEDLLQRIRT
Rv3291c|M.tuberculosis_H37Rv PSQPDDAPARLEHIEEVESCYSVAGEESYVLLVRVASARALEDLLQRIRT
Mflv_0397|M.gilvum_PYR-GCK -GNYKPLHSLLDVTPEVVEAHHVTGDDCFILKVLARSMSDLERLTGRLAT
MSMEG_3988|M.smegmatis_MC2_155 PADQERTVAALSTLSELRMCASITGDATIAFTVWTSSLADITRLERRIGE
MAV_3380|M.avium_104 -QHLTSVAEQIVRHDEVASVATVSGPANLMVVVICRDPEHFYRYLAERLA
. : *: ::* . * . : .
Mb3319c|M.bovis_AF2122/97 TA---NVRTRSTIILNTFYSDRQHIP------------
Rv3291c|M.tuberculosis_H37Rv TA---NVRTRSTIILNTFYSDRQHIP------------
Mflv_0397|M.gilvum_PYR-GCK LG---AVTTN--VVYSSPLTRRRLEPG-----------
MSMEG_3988|M.smegmatis_MC2_155 RLPWLSIRDSGVNLRTRKRMGWMLDPTGRATGVVVTPV
MAV_3380|M.avium_104 SVDHIQAYDVSVRSKRLKQVGSLIAHGRLVRAAR----