For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NEALDDIDRILVRELAADGRATLSELATRAGLSVSAVQSRVRRLESRGVVQGYSARINPEAVGHLLSAFV AITPLDPSQPDDAPARLEHIEEVESCYSVAGEESYVLLVRVASARALEDLLQRIRTTANVRTRSTIILNT FYSDRQHIP*
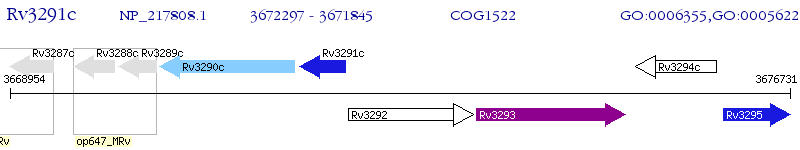
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3291c | - | - | 100% (150) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY ASNC-FAMILY) |
| M. tuberculosis H37Rv | Rv2324 | - | 1e-16 | 35.97% (139) | AsnC family transcriptional regulator |
| M. tuberculosis H37Rv | Rv2779c | - | 5e-10 | 30.28% (142) | LRP/AsnC family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3319c | - | 1e-78 | 99.33% (150) | AsnC family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_4797 | - | 7e-60 | 76.03% (146) | AsnC family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3647c | - | 1e-63 | 81.51% (146) | transcriptional regulatory protein |
| M. marinum M | MMAR_1242 | - | 7e-70 | 88.67% (150) | AsnC family transcriptional regulator |
| M. avium 104 | MAV_4263 | - | 2e-67 | 84.67% (150) | leucine-responsive regulatory protein |
| M. smegmatis MC2 155 | MSMEG_1763 | - | 1e-64 | 83.11% (148) | leucine-responsive regulatory protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2645 | - | 5e-70 | 88.67% (150) | AsnC family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1645 | - | 9e-61 | 78.38% (148) | AsnC family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Rv3291c|M.tuberculosis_H37Rv --------MNEALDDIDRILVRELAADGRATLSELATRAGLSVSAVQSRV
Mb3319c|M.bovis_AF2122/97 --------MNEALDDIDRILVRELAADGRVTLSELATRAGLSVSAVQSRV
MMAR_1242|M.marinum_M --------MGETLDDIDRILVRQLVSDGRATLAELAASAGLSVSAVQSRV
MUL_2645|M.ulcerans_Agy99 --------MGETLDDIDRILVRQLVSDGRATLAELAASAGLSVSAVQSRV
MAV_4263|M.avium_104 --------MSETLDDVDRILVRELVADGRATLAELAASARLSVSAVQSRV
MAB_3647c|M.abscessus_ATCC_199 MTVDSERTPERPLDDVDRILVRELARDGRATLAHLAAKAGLSISAVQTRV
MSMEG_1763|M.smegmatis_MC2_155 MRAMGEP-DEQSLDGIDRILVRELVADGRATLAHLAAAAGLSVSAVQARV
Mflv_4797|M.gilvum_PYR-GCK MTVPDDPQPPAPLDEIDRVLARELVADGRATLAHLAATAGLSVSAVQSRV
Mvan_1645|M.vanbaalenii_PYR-1 MTEPPDPQP---LDEIDRVLARELVADGRATLAHLAATAGLSVSAVQSRV
** :**:*.*:*. ***.**:.**: * **:****:**
Rv3291c|M.tuberculosis_H37Rv RRLESRGVVQGYSARINPEAVGHLLSAFVAITPLDPSQPDDAPARLEHIE
Mb3319c|M.bovis_AF2122/97 RRLESRGVVQGYSARINPEAVGHLLSAFVAITPLDPSQPDDAPARLEHIE
MMAR_1242|M.marinum_M RRLESRGVVTGYSARVDPEAVGHFLSAFVAITPLDPSQPDDAPARLEHIH
MUL_2645|M.ulcerans_Agy99 RRLESRGVVTGYSARVDPEAVGHFLSAFVAITPLDPSQPDDAPARLEHIH
MAV_4263|M.avium_104 RRLEARGVITGYSARIEPEAVGHRLSAFVAITPLDPSQPDDAPARLQHIE
MAB_3647c|M.abscessus_ATCC_199 RRLEARGVLAGYAARVNPESVGQLLSAFVAITPLDPSQPDDAPARLEHID
MSMEG_1763|M.smegmatis_MC2_155 RRLESRGVVTGYAARINPEALGNMLSAFVAITPLDPSQPDDAPARLEHIP
Mflv_4797|M.gilvum_PYR-GCK RRLESRGIVTGYTARIDPEALGSKLSAFVAITPLDPSQPDDAPARLEHIP
Mvan_1645|M.vanbaalenii_PYR-1 RRLESRGVVTGYTARIDPEAVGAKLSAFVAITPLDPSQPDDAPTRLEHIP
****:**:: **:**::**::* *******************:**:**
Rv3291c|M.tuberculosis_H37Rv EVESCYSVAGEESYVLLVRVASARALEDLLQRIRTTANVRTRSTIILNTF
Mb3319c|M.bovis_AF2122/97 EVESCYSVAGEESYVLLVRVASARALEDLLQRIRTTANVRTRSTIILNTF
MMAR_1242|M.marinum_M EVESCHSVAGEESYVLLVRVVSARALEDLLQRIRATANVRTRSTIILNTF
MUL_2645|M.ulcerans_Agy99 EVESCHSVAGEESYVLLVRVVSARALEDLLQRIRATANVRTRSTIILNTF
MAV_4263|M.avium_104 EIESCHSVAGEESYVLLVRVESARALEDLLQRIRTAANVRTRSTIILNTF
MAB_3647c|M.abscessus_ATCC_199 AIESCYSVAGEESYVLLVRVPSPRALEELLQLIRTTANVRTRSTIILQTF
MSMEG_1763|M.smegmatis_MC2_155 EIESCHSVAGDESYVLQVRVASARALEDLLQRIRTAANVRTRSTIILQTF
Mflv_4797|M.gilvum_PYR-GCK EIEACYSVAGEDNYMLIVHVESARALEGLLQKIRTAADVKTRSTIILHKF
Mvan_1645|M.vanbaalenii_PYR-1 EIEACYSVAGEDNYMLLVHVESARALEGLLQRIRTAADVKTRSTIILQKF
:*:*:****::.*:* *:* *.**** *** **::*:*:*******:.*
Rv3291c|M.tuberculosis_H37Rv YSDRQHIP-
Mb3319c|M.bovis_AF2122/97 YSDRQHIP-
MMAR_1242|M.marinum_M YSDRQFIP-
MUL_2645|M.ulcerans_Agy99 YSDRQFIP-
MAV_4263|M.avium_104 YDNRVYVP-
MAB_3647c|M.abscessus_ATCC_199 YDGRQLVP-
MSMEG_1763|M.smegmatis_MC2_155 YTGRDYIP-
Mflv_4797|M.gilvum_PYR-GCK YSERRYVPE
Mvan_1645|M.vanbaalenii_PYR-1 YSGRRHIPD
* * :*