For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVQSATEYGILVGVDSSAESDAAVRWAAREASLHDAPITLMHVIAPVVVSWPAGPYMATVLECQEENARH AIEQAQKVVADCLGETHGLTVQTEIRKESVARTLIDASKSAQMVVVGNRGMGALGRVLLGSTSTSLLHYA SGPVVVVHGDDQAAHDSRLPVLLGIDGSPASEVATSHAFDEASRRGVDLVALHVWIDVGDIPPIGPTWEE QEETGRALLAERLAGWQERYPDVKVHRRVERAQPAYWLLEEAKQAQLVVVGSHGRGGFTGMLLGSVSSRV AQSATTPVMVVRPR
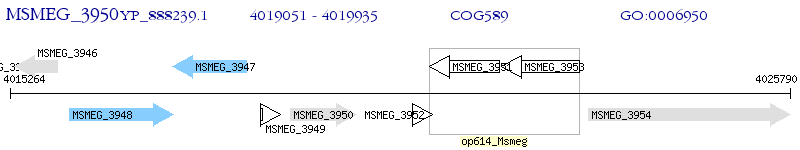
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3950 | - | - | 100% (294) | universal stress protein family protein |
| M. smegmatis MC2 155 | MSMEG_3940 | - | 2e-88 | 56.49% (285) | universal stress protein family protein |
| M. smegmatis MC2 155 | MSMEG_3936 | - | 3e-65 | 46.46% (297) | universal stress protein family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2051c | - | 5e-97 | 60.07% (293) | hypothetical protein Mb2051c |
| M. gilvum PYR-GCK | Mflv_4992 | - | 2e-85 | 53.08% (292) | UspA domain-containing protein |
| M. tuberculosis H37Rv | Rv2026c | - | 5e-97 | 60.07% (293) | hypothetical protein Rv2026c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2489 | - | 3e-40 | 37.54% (301) | hypothetical protein MAB_2489 |
| M. marinum M | MMAR_2995 | - | 3e-95 | 59.73% (293) | hypothetical protein MMAR_2995 |
| M. avium 104 | MAV_2506 | - | 2e-88 | 59.09% (286) | universal stress protein family protein |
| M. thermoresistible (build 8) | TH_2494 | - | 2e-67 | 46.95% (311) | PUTATIVE putative conserved hypothetical membrane protein |
| M. ulcerans Agy99 | MUL_2236 | - | 3e-94 | 59.04% (293) | hypothetical protein MUL_2236 |
| M. vanbaalenii PYR-1 | Mvan_1415 | - | 1e-85 | 55.07% (296) | UspA domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3950|M.smegmatis_MC2_155 MVQSATEYGILVGVDSSAESDAAVRWAAREASLHDAPITLMHVIAPVVVS
Mvan_1415|M.vanbaalenii_PYR-1 MPGETTRYGIVVGVDGSPESSAALQWAGGEALLFREPVTLIYAVAPTVVS
Mb2051c|M.bovis_AF2122/97 MSAATAKYGILVGVDGSAQSNAAVAWAAREAVMRQLPITLLHIVAPVVVG
Rv2026c|M.tuberculosis_H37Rv MSAATAKYGILVGVDGSAQSNAAVAWAAREAVMRQLPITLLHIVAPVVVG
MMAR_2995|M.marinum_M MSSGAKKYGILVAVDGSAQSDAAVAWAAREAGLHRTPITLMHGVAPVVVG
MUL_2236|M.ulcerans_Agy99 MSSGAKKYGILVAVDGSAQSDAAVPWAAREAGLHRTPITLMHGVAPVVVG
MAV_2506|M.avium_104 MSD-PKHRGILVCVDGSAASDAAVAWAAREAAMRGLPITVIHAVAPVVVG
Mflv_4992|M.gilvum_PYR-GCK MSDPSTDLGILVGVDGSPESHAAVRWAAQEAVLRRRPVTLMHVVSPIVVT
TH_2494|M.thermoresistible__bu VSSTAAYEGIVVGVDGSAASADAVRWAARSAALRNLQLTLVHVMTATSIG
MAB_2489|M.abscessus_ATCC_1997 MTTSTPNSHILVGVDGSTSALHALTWAGAEAQRRNLPLTLVHVIDHSSLG
: . *:* **.*. : *: **. .* :*::: : :
MSMEG_3950|M.smegmatis_MC2_155 ----WPAGPYMATVLECQEENARHAIEQAQKVVADCLGET----------
Mvan_1415|M.vanbaalenii_PYR-1 ----WPMAPLQDTVVECERQNAEEALQYARNAVASVAESCG---------
Mb2051c|M.bovis_AF2122/97 ----WPVGQLYANMTEWQKDNAQQVIEQAREALTNSLGES----------
Rv2026c|M.tuberculosis_H37Rv ----WPVGQLYANMTEWQKDNAQQVIEQAREALTNSLGES----------
MMAR_2995|M.marinum_M ----WPVGQLYAEMPDWQRDEANSVMERARKVLSEALGDS----------
MUL_2236|M.ulcerans_Agy99 ----WPVGQLYAEMPDWQRDEANSVMERARKVLSEALGDS----------
MAV_2506|M.avium_104 ----WPVGQLYADMPAWQQDGAQQVIDQARKIVIANQGGG----------
Mflv_4992|M.gilvum_PYR-GCK ----WPIDTVVANFYEWQEDNAARVLKQSQETLAAAVSDT----------
TH_2494|M.thermoresistible__bu APLVWPAGPIPDSLIQWQEDEARQILADSAEIAENAFQEARAAGPGPAGD
MAB_2489|M.abscessus_ATCC_1997 QG--YNLG-ASASFFQHLETDGAEFLSQAKDHVYALHPNV----------
: . . . : : .
MSMEG_3950|M.smegmatis_MC2_155 HGLTVQTEIRKESVARTLIDASKSAQMVVVGNRGMGALGRVLLGSTSTSL
Mvan_1415|M.vanbaalenii_PYR-1 QDIDIRTEVRYGPPVQVLVDASRQARMVVVGNRGRGALGRLALGSVSDGL
Mb2051c|M.bovis_AF2122/97 KPPQVHTELVFSNVVPTLIDASQQAWLMVVGSQGMGALGRLLLGSISTAL
Rv2026c|M.tuberculosis_H37Rv KPPQVHTELVFSNVVPTLIDASQQAWLMVVGSQGMGALGRLLLGSISTAL
MMAR_2995|M.marinum_M ESPDVRTEVVYSNVVAALIEASTQASMVVVGSQGMGALGRMLLGSVCSAL
MUL_2236|M.ulcerans_Agy99 ESPDVRTEVVYSNVVAALIEASTQASMVVVGSQGMGALGRMLLGSVCSAL
MAV_2506|M.avium_104 TPPEMRTEIIYSAVTPTLIDASRDAWMIVAGSQGLGALGRLLLGSVTAAL
Mflv_4992|M.gilvum_PYR-GCK TAPTVEVEVRHDGIVPEFTEASQHADLLVLGSRGLGPVGGAVLGSVSRAL
TH_2494|M.thermoresistible__bu DAVRVVTGVVRAAPVPTLVDASKQAKLVAVGSRGQGPLRRVLLGSVSSGL
MAB_2489|M.abscessus_ATCC_1997 ---EVSTVKATGRPVPVLVELSRNALMTVLGSSGLGGFTGMMAGSVSVSL
: . . : : * * : . *. * * . ** .*
MSMEG_3950|M.smegmatis_MC2_155 LHYASGPVVVVHGDDQAAHD-SRLPVLLGIDGSPASEVATSHAFDEASRR
Mvan_1415|M.vanbaalenii_PYR-1 IHHAHGAVTVVHTRRGHLPD-PAASVLLGVDGSPASEDATALAFEEAERR
Mb2051c|M.bovis_AF2122/97 LHHARCPVAIIHSGNGATPD-SDAPVLVGIDGSPASEAATALAFDEASRR
Rv2026c|M.tuberculosis_H37Rv LHHARCPVAIIHSGNGATPD-SDAPVLVGIDGSPASEAATALAFDEASRR
MMAR_2995|M.marinum_M LHHAHCPVAVVHSDSGAAPD-PGAAVVVGIDGSPAAEAATAMAFEEASRR
MUL_2236|M.ulcerans_Agy99 LHHAHCPVAVVHSDSGAAPD-PGAAVVVGIDGSPAAEAATAMAFEEASRR
MAV_2506|M.avium_104 LAHAHCPVAVVHTDDHAARA-ADAPVLVGTDGSPASEAAIALAFDEASRR
Mflv_4992|M.gilvum_PYR-GCK LHHAQCPVVIAKEGVVPRDA-GALPVLLGIDGSPASENATTFAFDEASRR
TH_2494|M.thermoresistible__bu VHHAHCPVAVIREGTPHTPDFAKRPVLVGIDGSESAQRATAVAFEEASFR
MAB_2489|M.abscessus_ATCC_1997 TAKGHSPVVVVRES--PVPG--GGPVVVGIDDSESSDGAAAWAFEEAAMR
. .*.: : .*::* *.* ::: * : **:** *
MSMEG_3950|M.smegmatis_MC2_155 GVDLVALHVWIDVG------DIPPIGPTWEEQEETGRALLAERLAGWQER
Mvan_1415|M.vanbaalenii_PYR-1 GVDLVAVHVWGDVG------GLPLQGDEWRGHRQQAEEVLAERLAGWQER
Mb2051c|M.bovis_AF2122/97 RVDLVALHAWTDLG------MFPVLGMDWREREKREAEVLAERLAGWQEQ
Rv2026c|M.tuberculosis_H37Rv RVDLVALHAWTDLG------MFPVLGMDWREREKREAEVLAERLAGWQEQ
MMAR_2995|M.marinum_M GVDLVALHAWSDVG------VFPILGMDWRERESQGEEILAERLAGWQEQ
MUL_2236|M.ulcerans_Agy99 GVDLVALHAWSDVG------VFPILGMDWRERESQGEEILAERLASWQQQ
MAV_2506|M.avium_104 GVGVVALHAWSDVG------VFPILGMDWRDSEAKGEELLAERLAGWQEQ
Mflv_4992|M.gilvum_PYR-GCK GADLVALHAWSDVA------VFPILGMDWHQYEQQGHEILAERLAGWQER
TH_2494|M.thermoresistible__bu GVDLIALHAWSDAD------IADIPTLELSAVQQSAERVLSERLAGFRER
MAB_2489|M.abscessus_ATCC_1997 ETELVAVHVWNNIPPGYVYAYTAWSAMDSAGEQQRQEQLLAERLAGWQEK
. ::*:*.* : . :*:****.::::
MSMEG_3950|M.smegmatis_MC2_155 YPDVKVHRRVERAQPAYWLLEEAKQAQLVVVGSHGRGGFTGMLLGSVSSR
Mvan_1415|M.vanbaalenii_PYR-1 HPDVRVSRHVEFDDPAHHLVDRARTAQLVVLGSHGRGGFAGMLLGSVSST
Mb2051c|M.bovis_AF2122/97 YPDVRVHRSLVCDKPARWLLEHSEQAQLVVVGSHGRGGFSGMLLGSVSSA
Rv2026c|M.tuberculosis_H37Rv YPDVRVHRSLVCDKPARWLLEHSEQAQLVVVGSHGRGGFSGMLLGSVSSA
MMAR_2995|M.marinum_M YPDVHVKRSLVCDKPAHWLLEEAQHAQLVIVGNRGRGGFPGMLLGSVSST
MUL_2236|M.ulcerans_Agy99 YPDVHVKRSLVCDKPAHWLLEEAQHAQLVIVGNRGRGGFPGMLLGSVSST
MAV_2506|M.avium_104 YPDVHVKRLVVCDKPSRWLVAEAERAQLVVVGSHGRGGFPGMLLGSVSSH
Mflv_4992|M.gilvum_PYR-GCK YPDVHIRRRIVCDTPARWLIDESRHSQLVVVGSRGRGGIAGLLLGSVSTT
TH_2494|M.thermoresistible__bu YPDVSVHRRIVGSEPARHLVEASEDAQLVVVGSRGRGGFTGMLLGSVSRA
MAB_2489|M.abscessus_ATCC_1997 YPDVPVRRVVAVGHPADVLLSQAAGAQLIVTGSRGRGDIAGFFLGSTSHA
:*** : * : *: *: : :**:: *.:***.:.*::***.*
MSMEG_3950|M.smegmatis_MC2_155 VAQSATTPVMVVRPR
Mvan_1415|M.vanbaalenii_PYR-1 VARSVDIPVTVARPR
Mb2051c|M.bovis_AF2122/97 VAHSVRIPVIVVRPS
Rv2026c|M.tuberculosis_H37Rv VAHSVRIPVIVVRPS
MMAR_2995|M.marinum_M VAHSAKVPVIVVRPA
MUL_2236|M.ulcerans_Agy99 VAHSAKVPVIVVRPA
MAV_2506|M.avium_104 VAQSATAPVIVVRGR
Mflv_4992|M.gilvum_PYR-GCK VAESALAPVAVVRG-
TH_2494|M.thermoresistible__bu VVNAVTTPVIVAR--
MAB_2489|M.abscessus_ATCC_1997 LIHKASCPVLVAPRT
: . . ** *.